Introduction
GazooResearch helps medical researchers (like yourself) cultivate massive medical databases. You’ll find that GazooResearch has tools for every step of the research process from creating and using standardized data dictionaries, clinical information extraction, reviewing patient data for accuracy, data analysis, and it even provides data for the development of artificial intelligence algorithms.
Figue 1. GazooResearch Overview
Installation
Requirements
- Read/Write and Internet priviliges!
- MacOS 13/14/15 or Windows 10/11
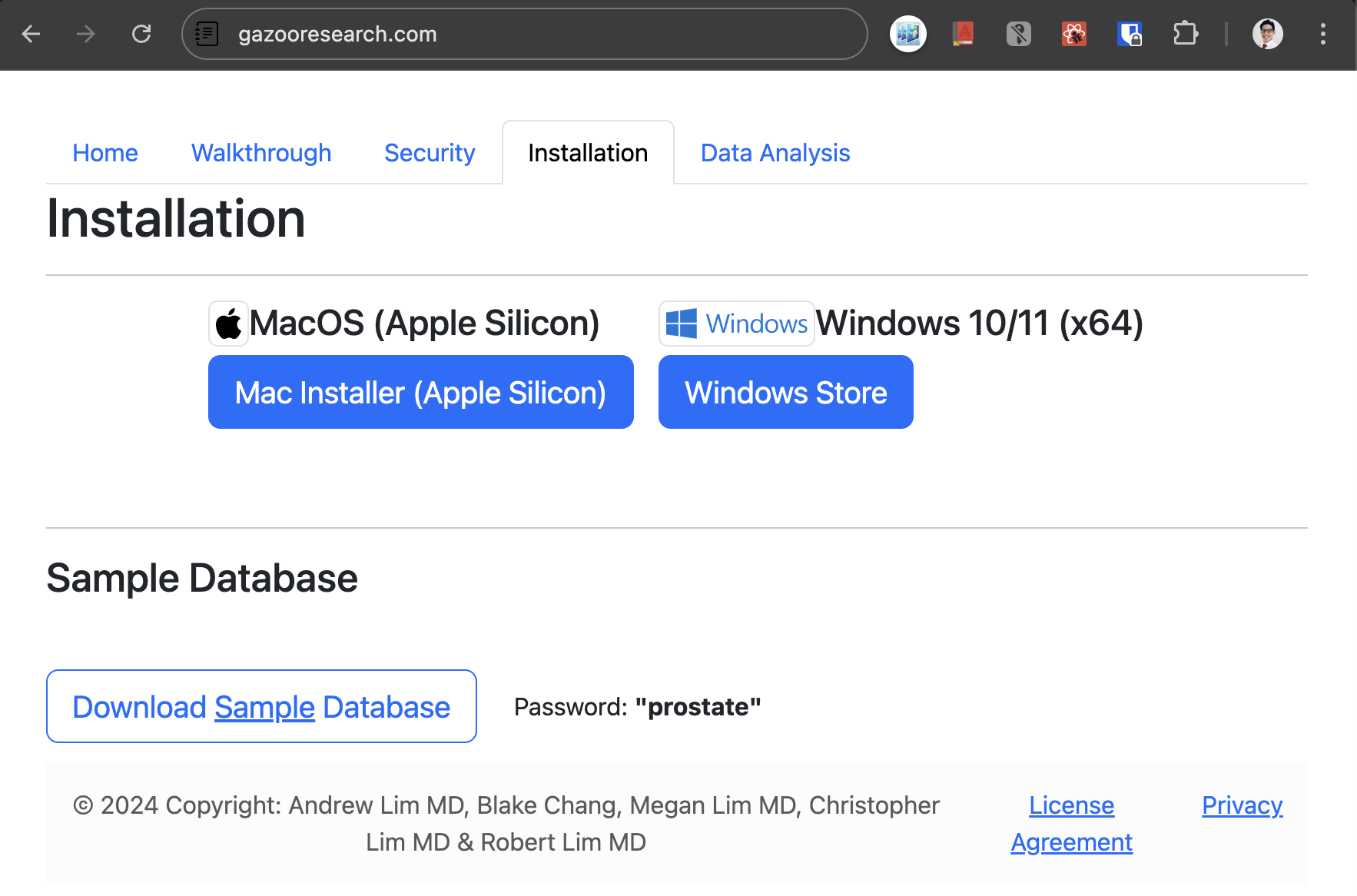
Installation is as simple as navigating to the GazooResearch website https://gazooresearch.com, and clicking on the MacOS download button for the installable .dmg file, or navigating to the windows store to download the application.
Figure 1. Website Installation Page
Creating A Database
A gazoo databases is one simple file with a suffix “.gazoo”. This is akin to an excel database file ending in “.xlsx”. You can save this encrypted database file on your local computer, or to your organization’s encrypted OneDrive.
To create a new database follow these steps:
- Open GazooResearch.app
- Click: “New Database” and select the location and name of the database.
- Set and Confirm the database’s password.
- Click: “Create Database” button
You should see a new file named: “<database name>.gazoo”.
Figure 1. Create A Database
Uploading Documents
You can upload documents via the Upload Documents tab.
Click on “Choose Files”
You can upload multiple documents at once. The documents must be either pdf, jpeg, or png.
During the uploading process the documents get encrypted and saved in the .gazoo file.
You can view the uploaded documents in the “Review Documents” Tab.
Figure 2. Uploading Documents
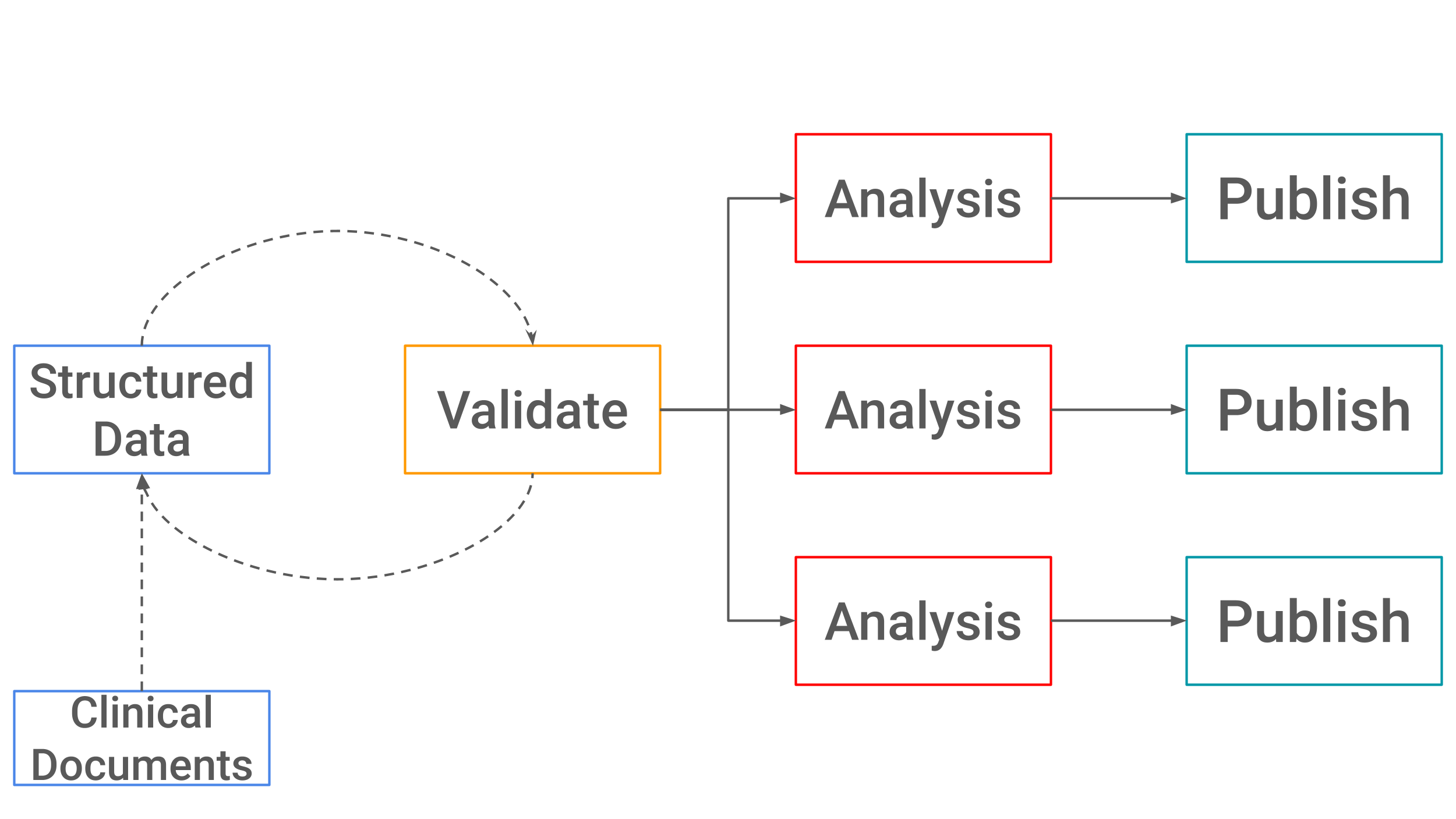
Data Structure
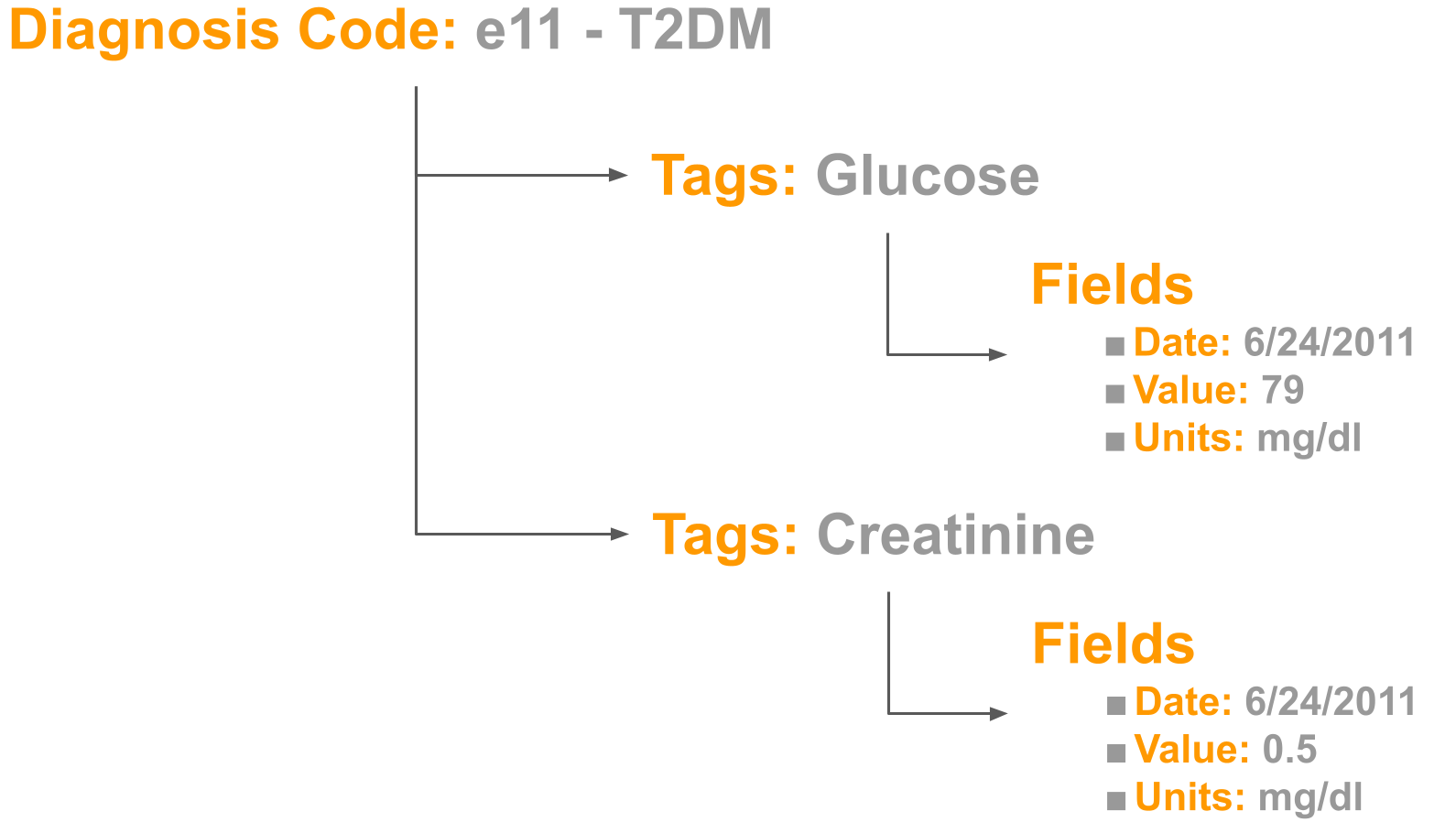
GazooResearch’s data structure has 4 layers. 1) Diagnosis, 2) Tag, 3) Field, and 4) Value.
Let’s illustrate why 4 layers are needed with an example. Say we are wanting to study the correlation of blood sugars, and kidney function in patients with diabetes. We want to collect serum glucose levels, and serum creatinine levels (Figure 1.)
Figure 1. Lab Result
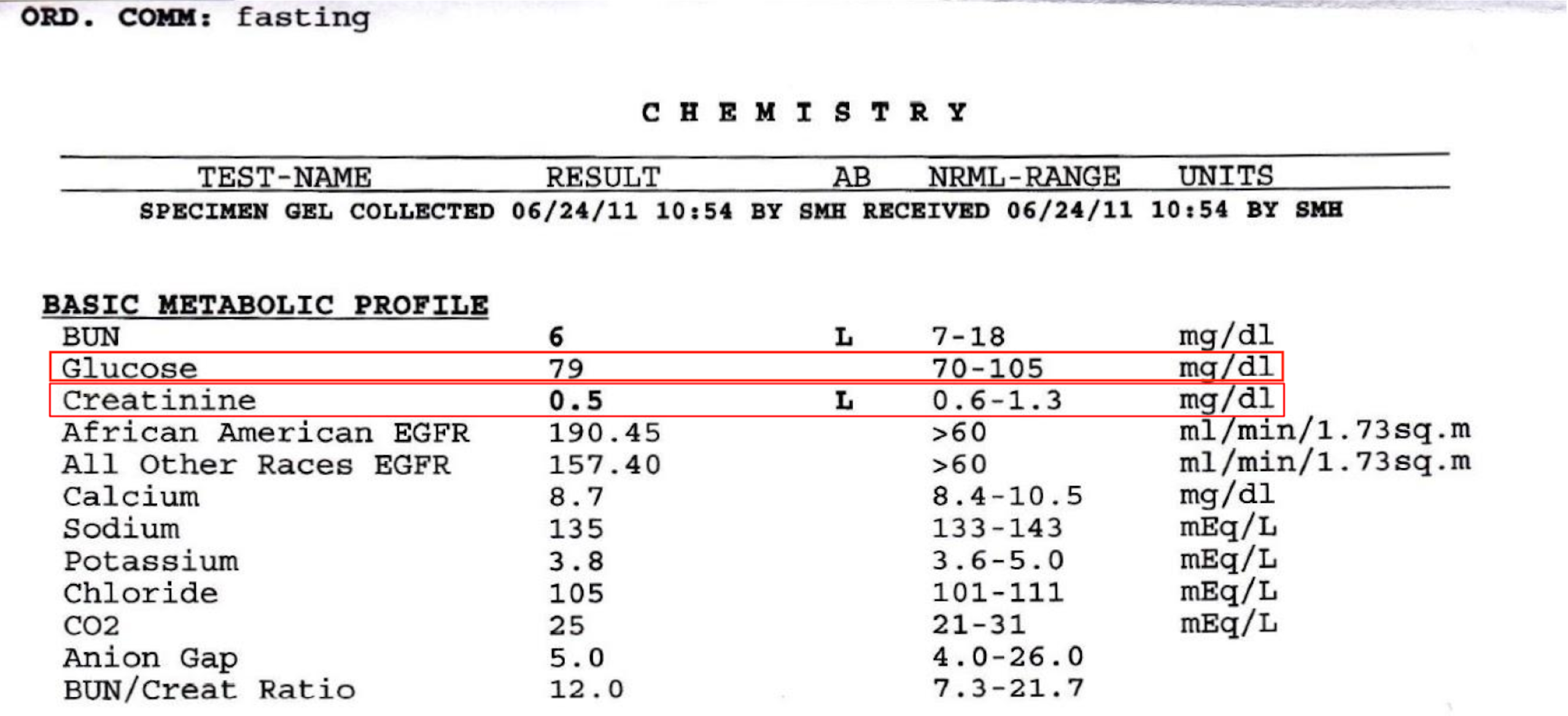
At first glance, you might think you’ll just need to store one number for glucose and another creatinine: 79 for glucose, and 0.5 for creatinine. Unfortunately, this alone is not enough information for your analysis. In reality, Glucose actually has 3 fields: 1) Date, 2) Value, and 3) Units. Creatinine also has 3 fields.
Finally, since we are studying diabetes we also have a base layer which is the icd10 code for diabetes.
Figure 2. GazooResearch’s Data Structure
When defining data structures in GazooResearch, we allow for the 4 layers as shown in the video below.
Figure 3. Data Dictionary
Data Analysis
Within the data analysis python modules you will also see this exact 4 layer data structure.
{'icd10':str, 'tag': str, 'field':str, 'value': [str, str, ...]},
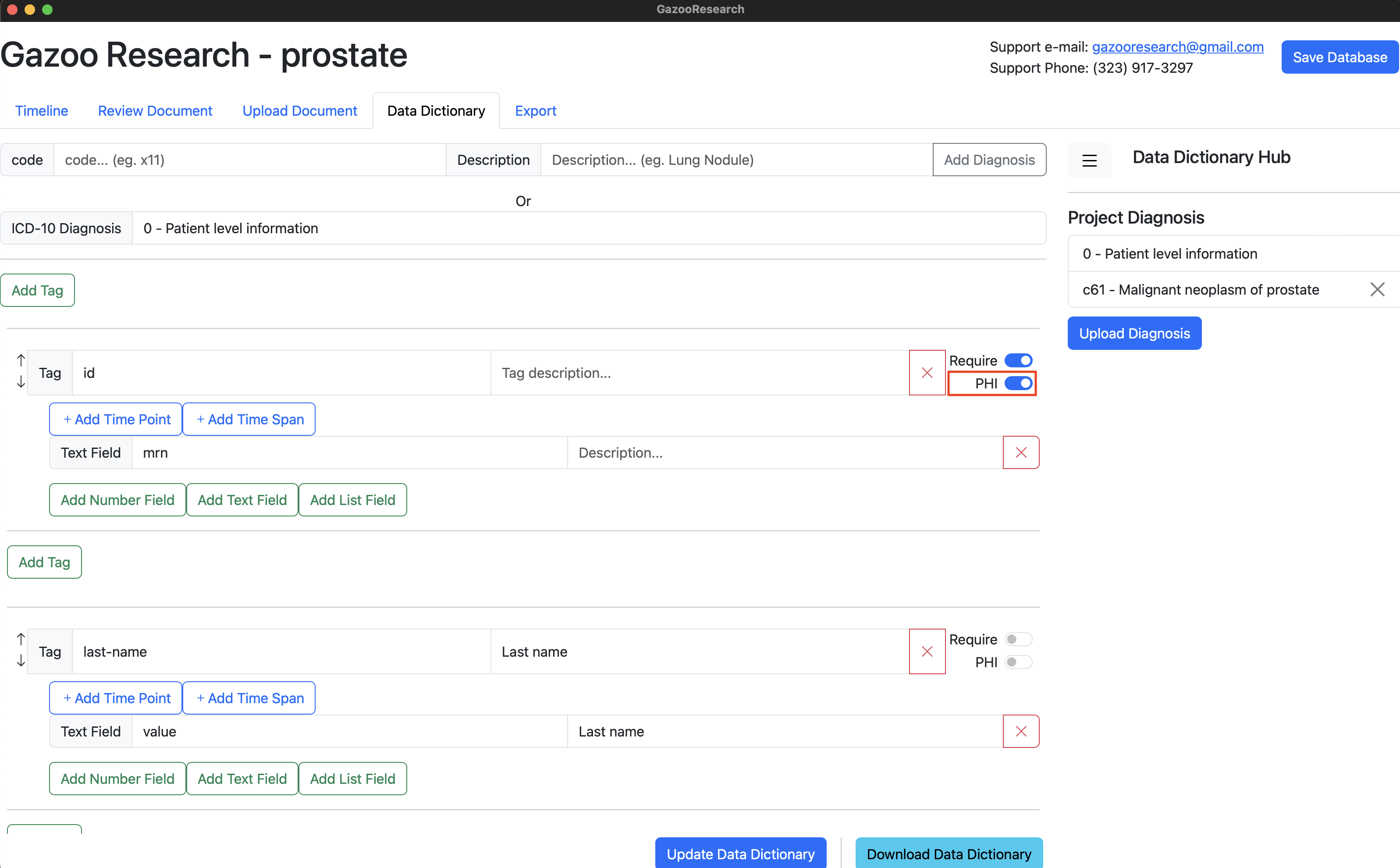
Data Dictionary
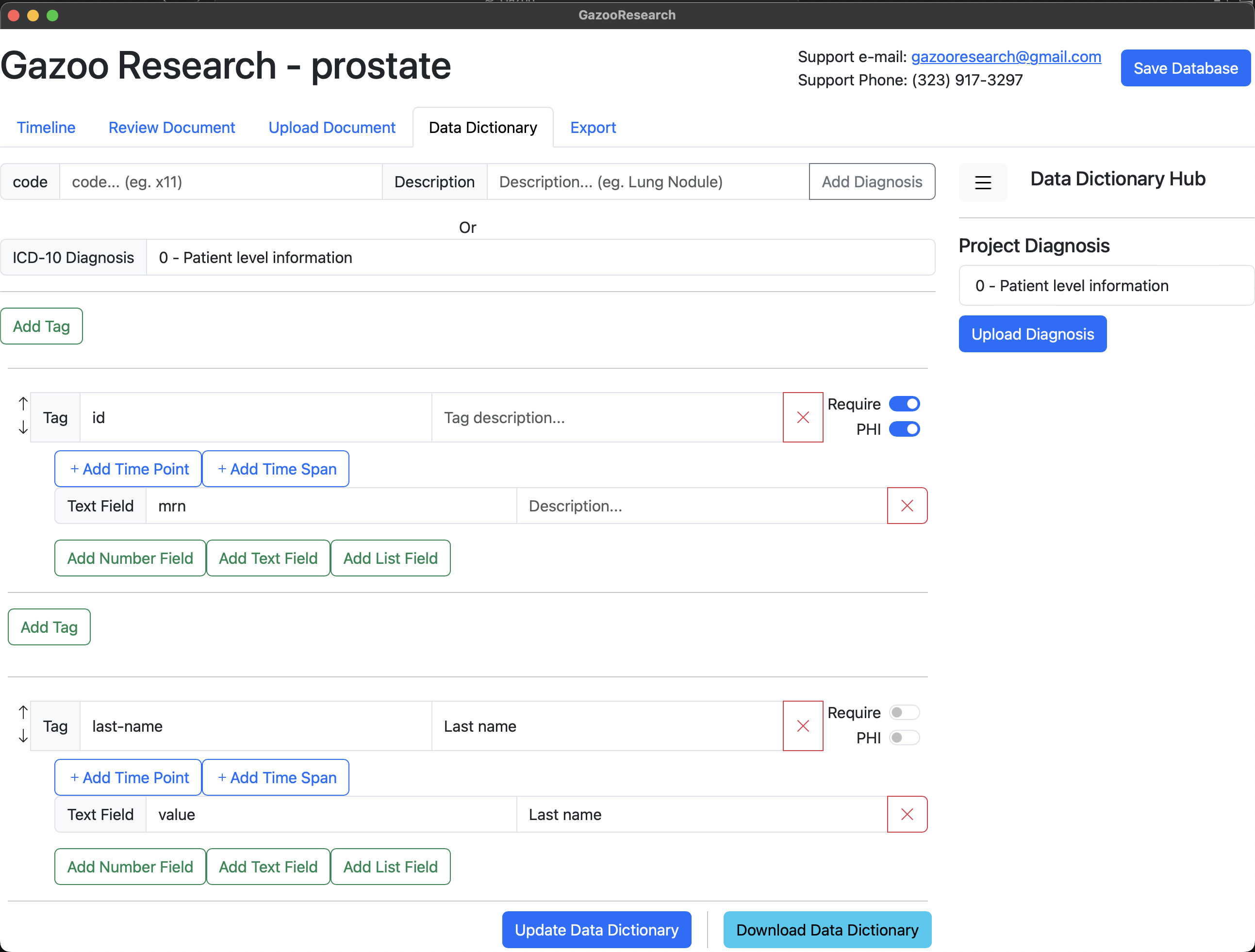
Your database has a basic “Patient Level Information” data dictionary already loaded. This has information like patient “id”, “last-name”, “first-name”, “dob”, “sex”, “document”, “death”.
Figure 1. Patient Level Information
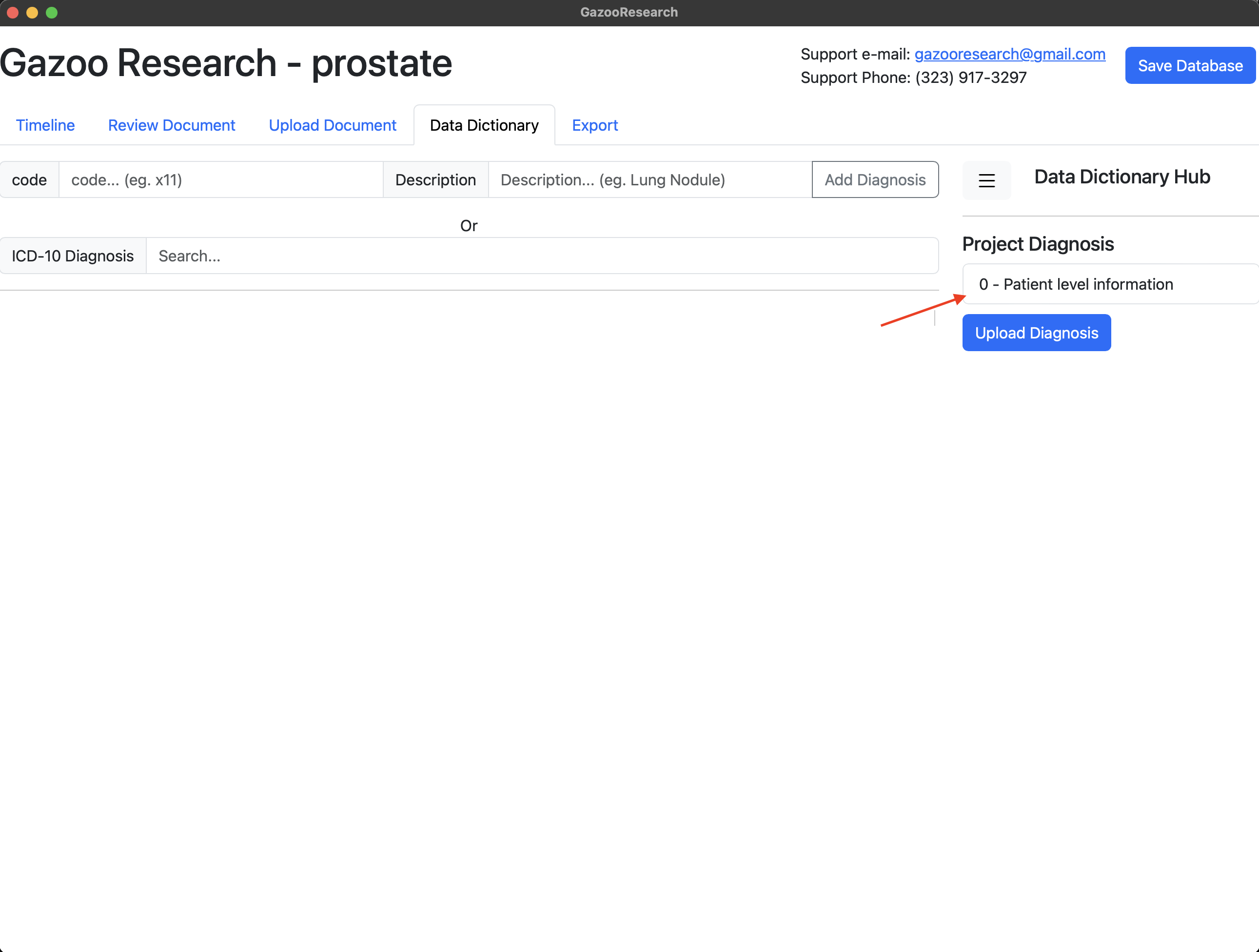
The information that is relavant to the patient
Creating a Data Dictionary
You can either download a pre-made data dictionary from the Data Dictionary Hub, or create a custom data dictionary to fit your exact needs.
Data Dictionary Hub
GazooResaerch will host a variety of premade data dictionaries.
Custom Data Dictionary
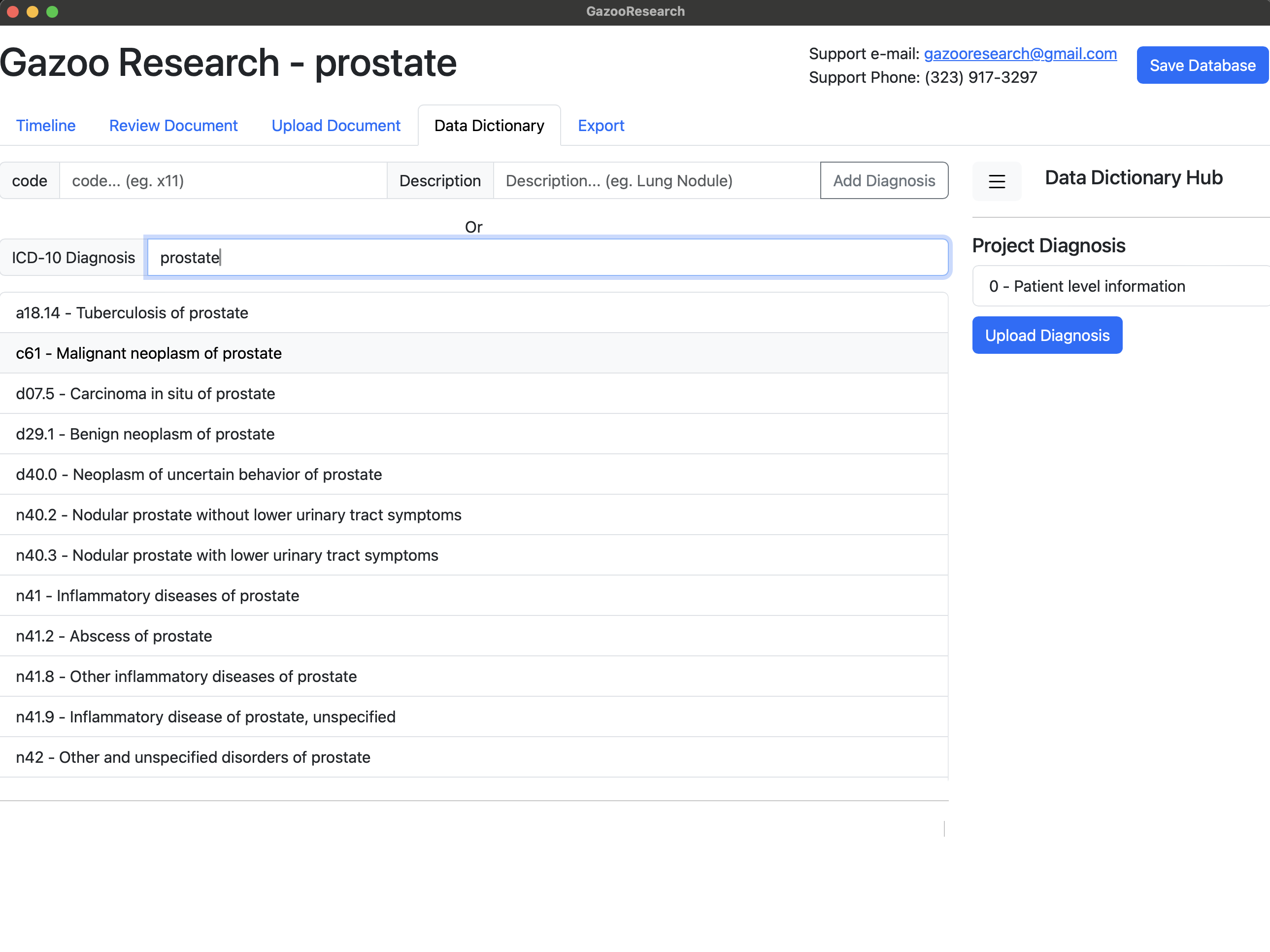
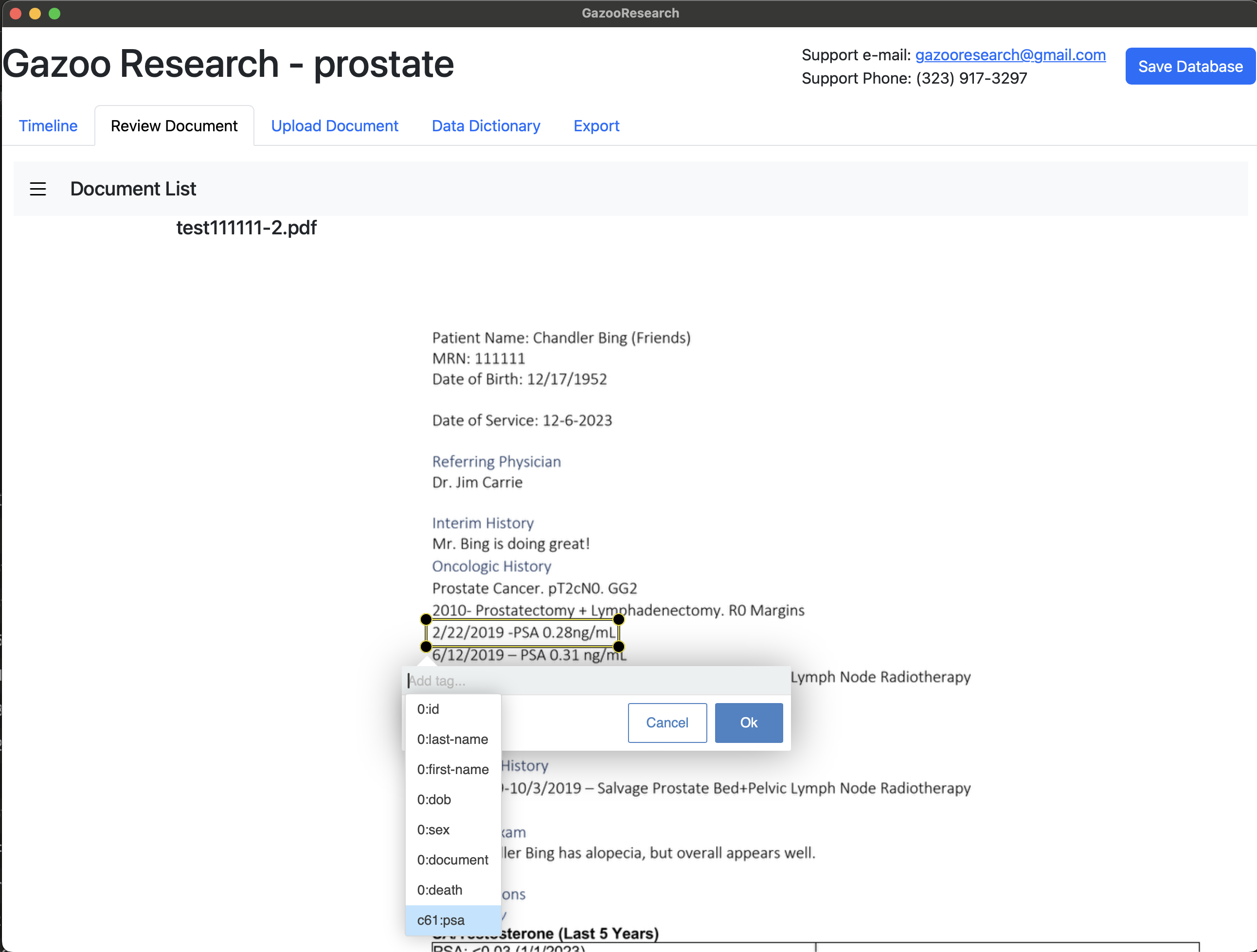
In this example, we are going to create a data dictionary to study prostate cancer. Firstly, search and select the icd-10 diagnosis for prostate cancer.
We will want to extract PSA values. So lets create a tag for PSA.
 For each tag you can add a description, so that other people will understand what the acronym ‘psa’ means.
For each tag you can add a description, so that other people will understand what the acronym ‘psa’ means.
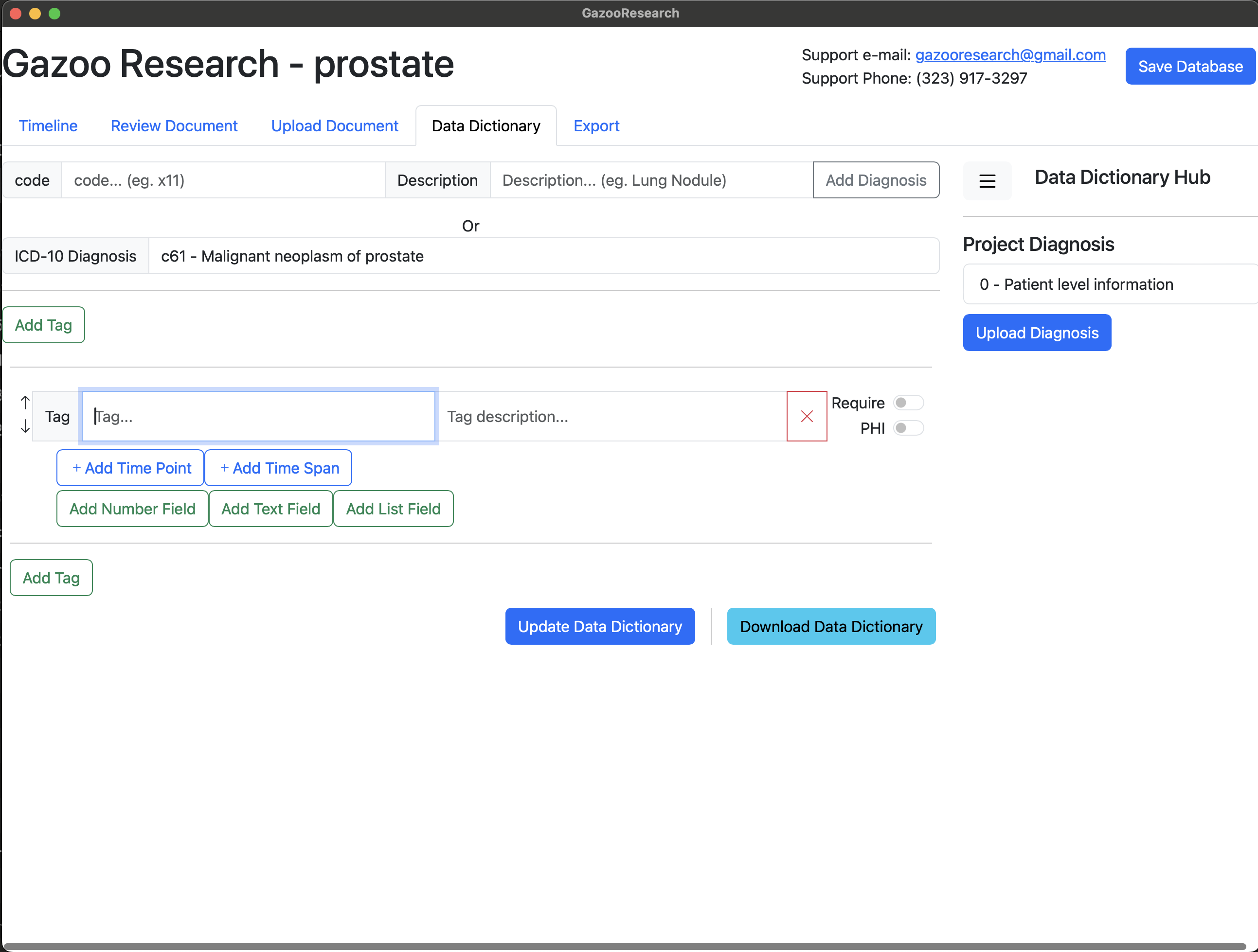
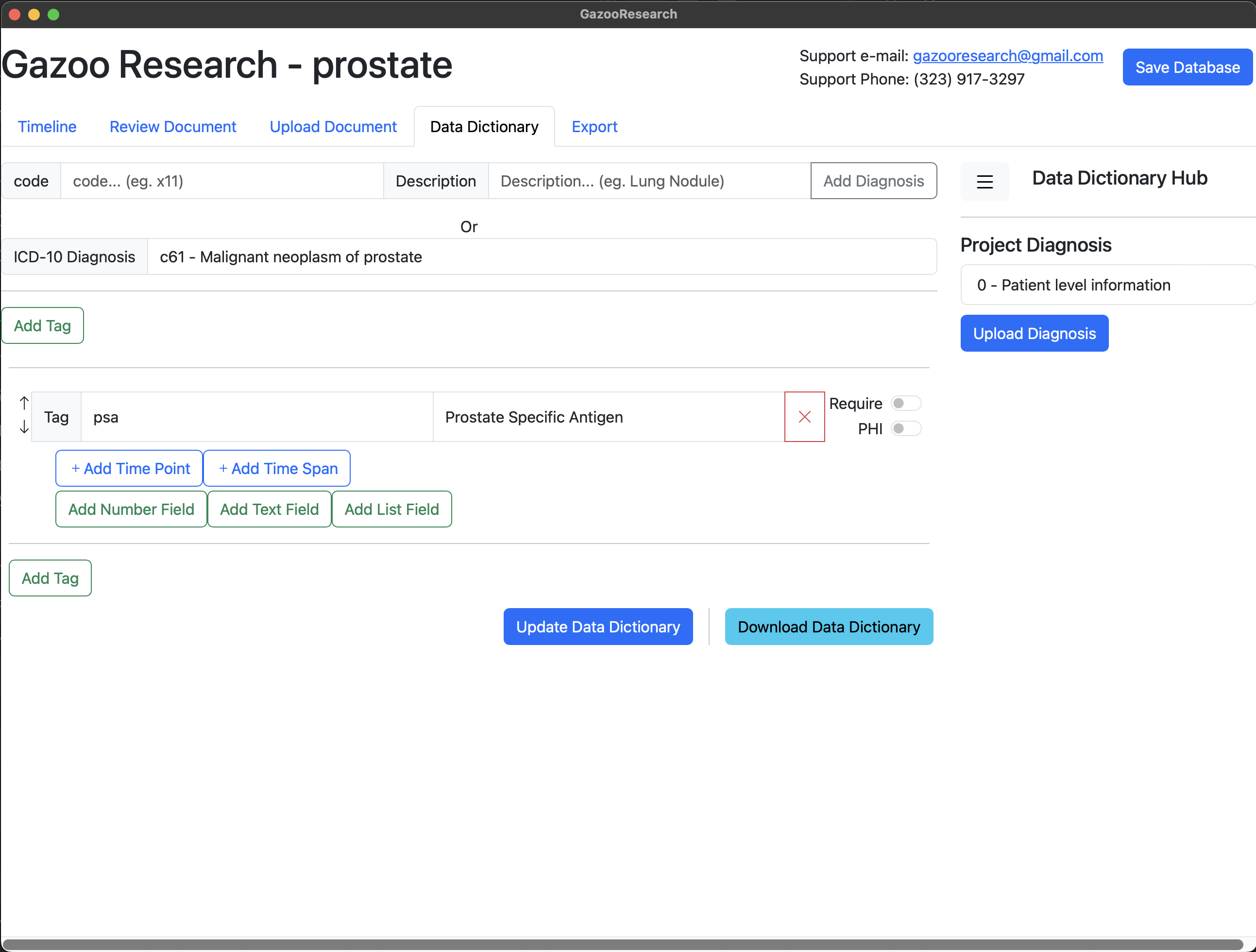 Next we want to add the fields for PSA. In the case of PSA the fields include 1) Date, 2) Value, and 3) Units. So first we will add the timepoint.
Next we want to add the fields for PSA. In the case of PSA the fields include 1) Date, 2) Value, and 3) Units. So first we will add the timepoint.
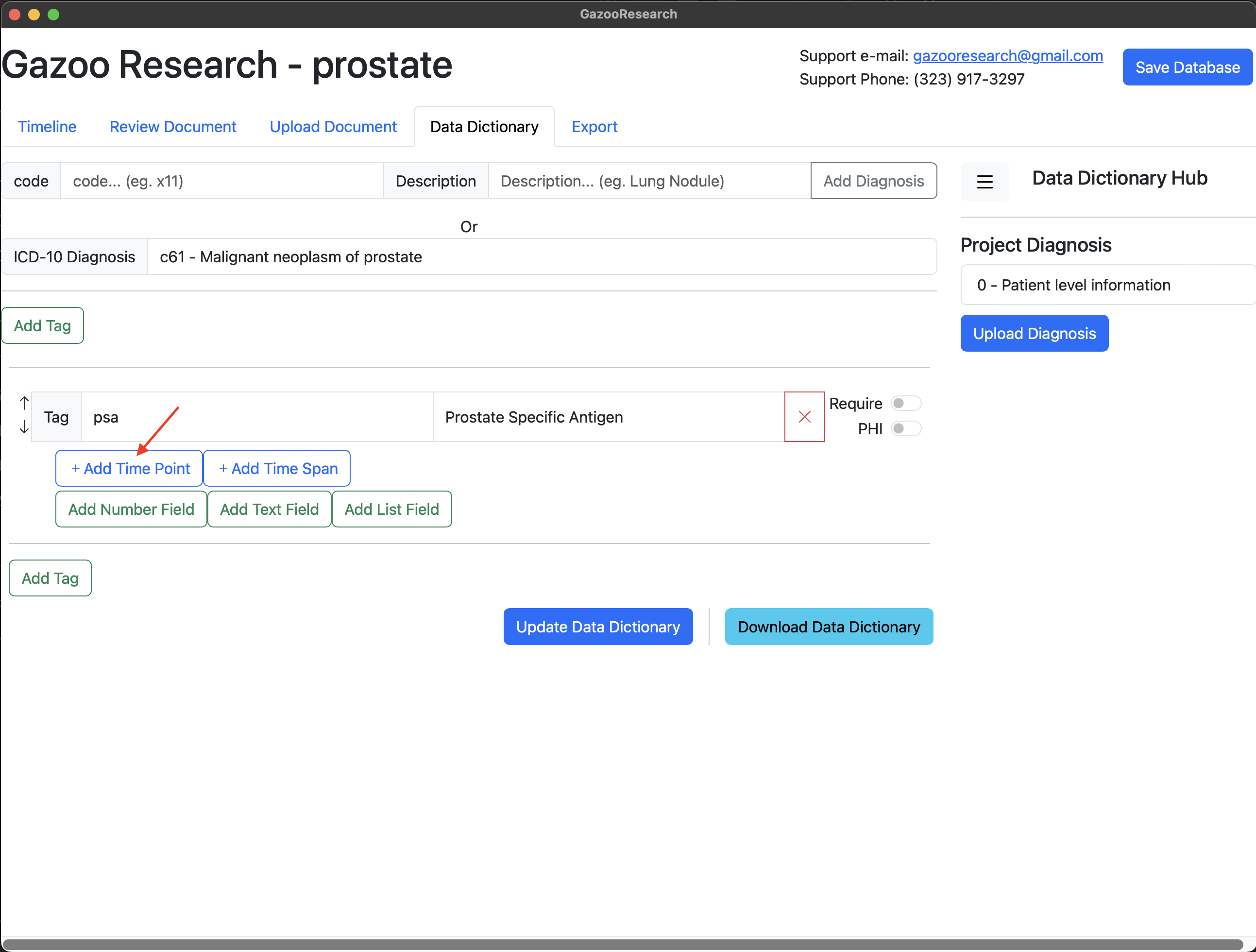
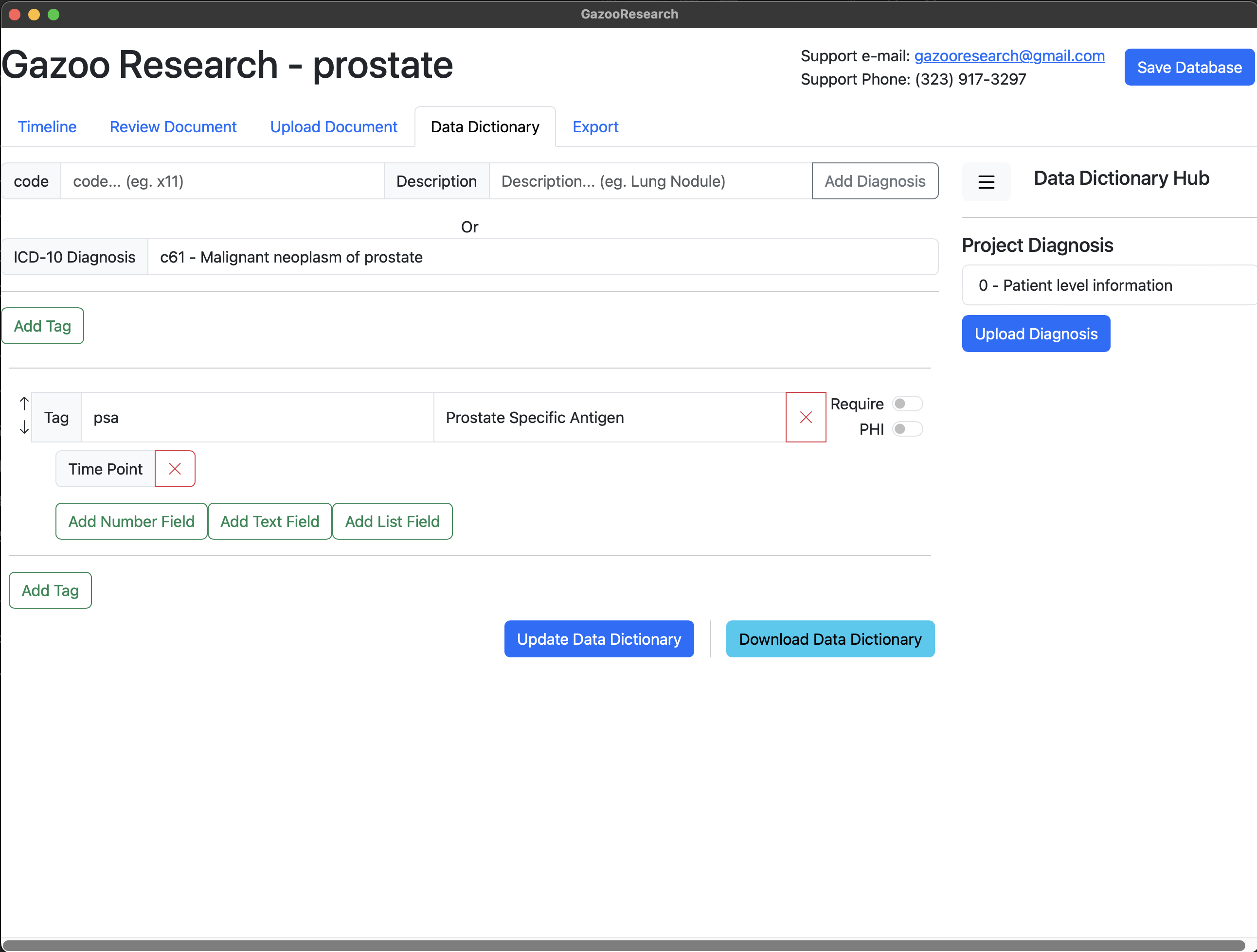 Lastly we’ll add the units field. Note that different hospital may report PSA using different units (ng/ml, g/dL). Therefore units is a categorial variable, and can have one of two values, therefore we will use the “List Field” option.
Lastly we’ll add the units field. Note that different hospital may report PSA using different units (ng/ml, g/dL). Therefore units is a categorial variable, and can have one of two values, therefore we will use the “List Field” option.
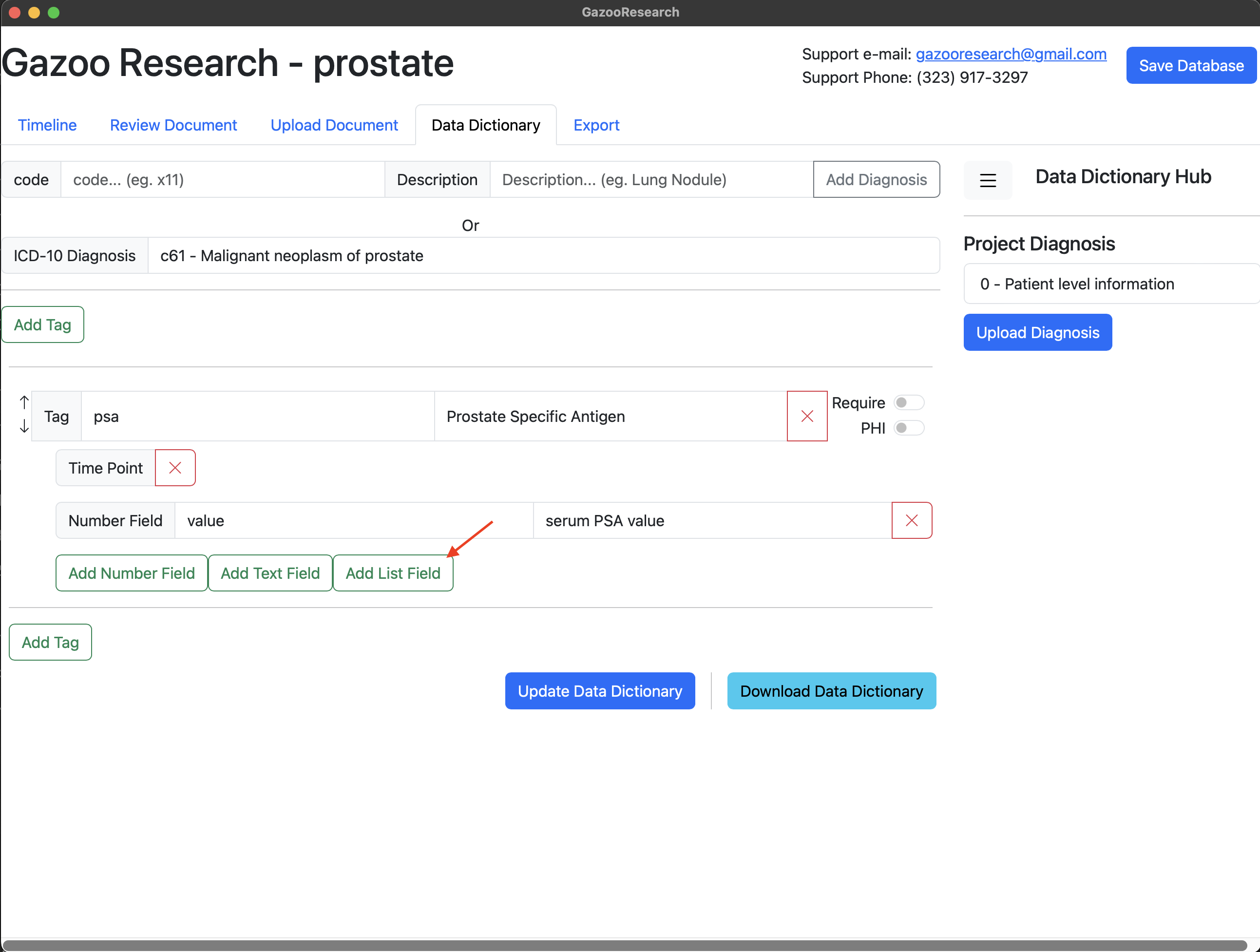
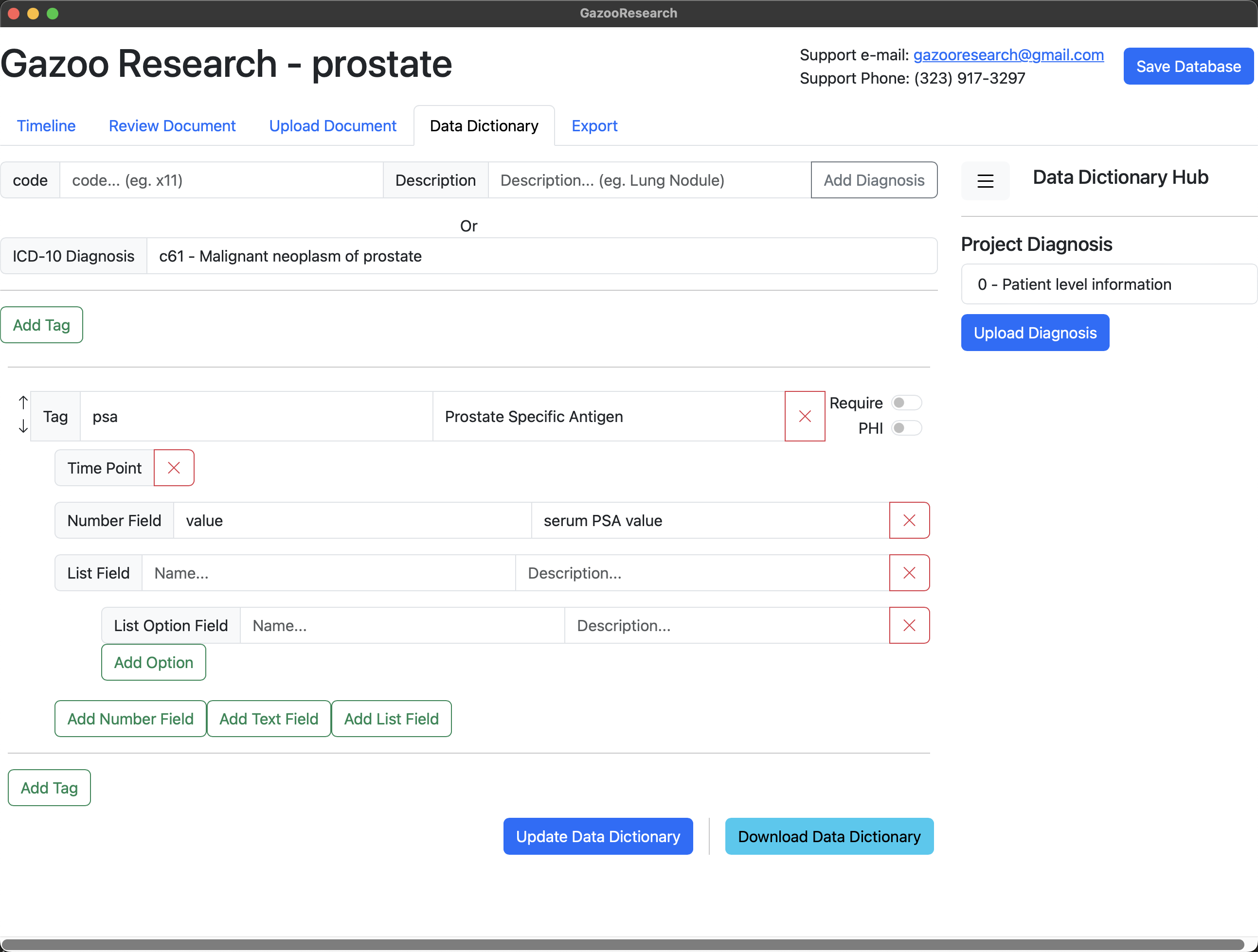 We name the list “units”, and give it two possible values. Either ng/ml or g/dL
We name the list “units”, and give it two possible values. Either ng/ml or g/dL
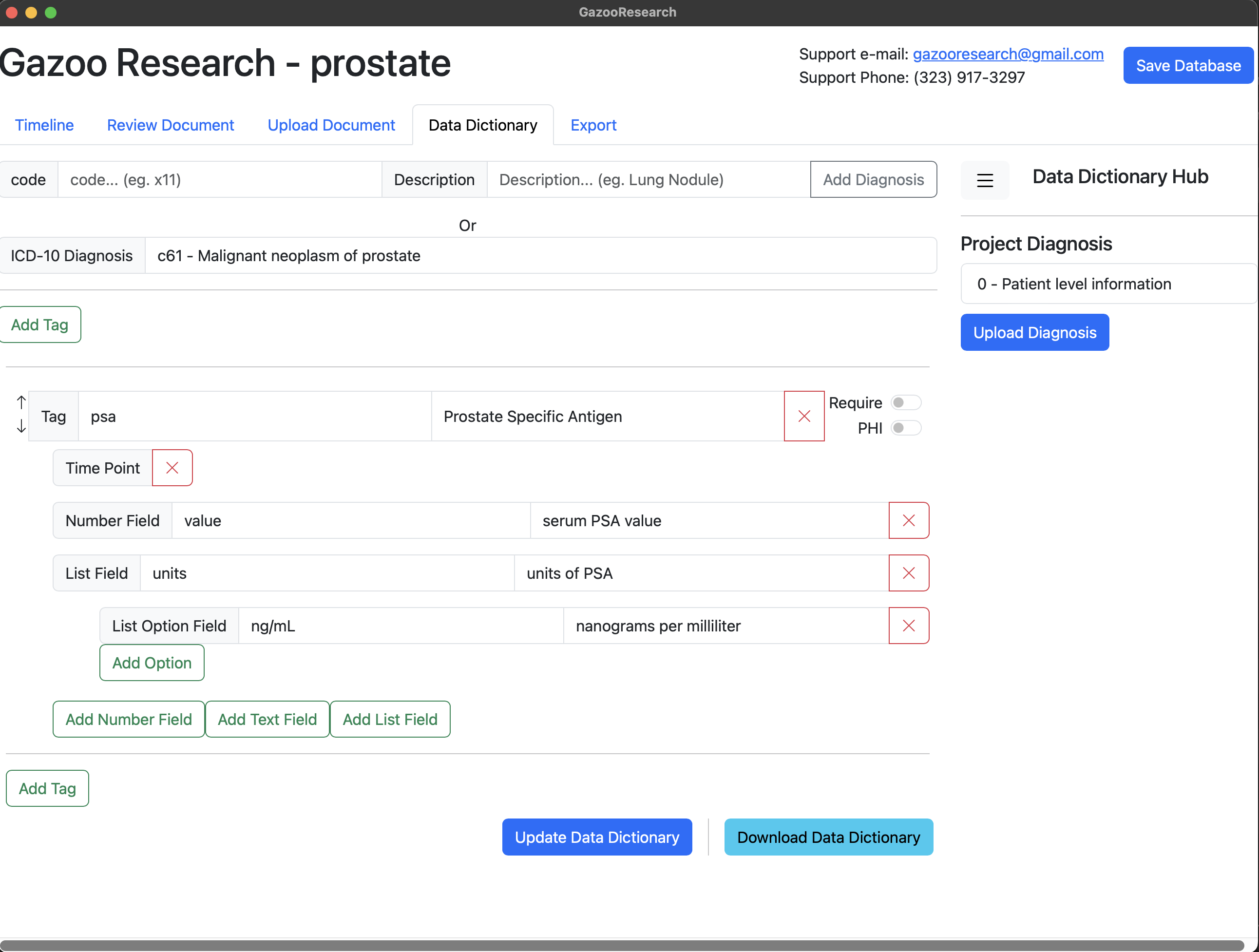
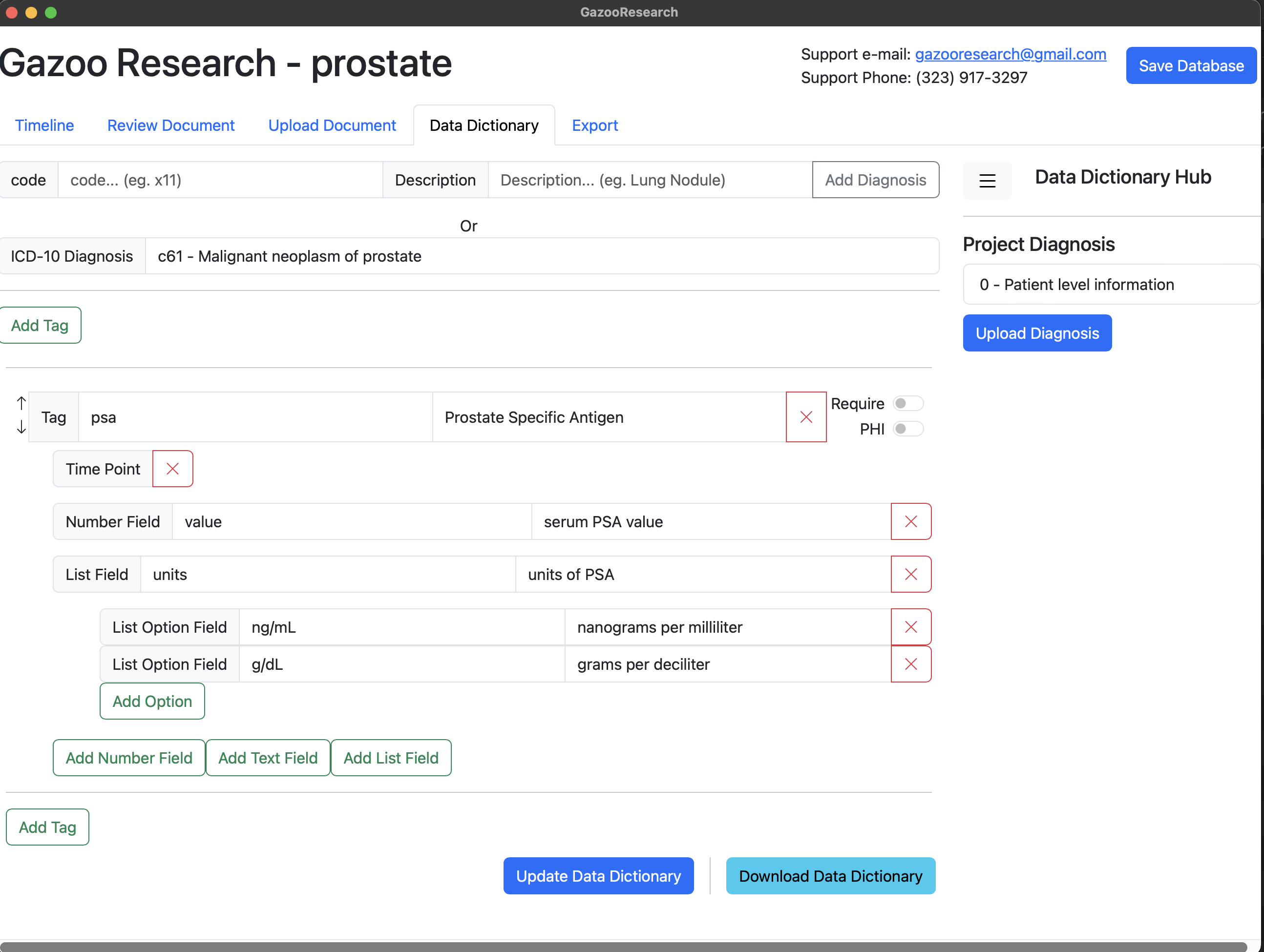 Finally lets save the data dictionary.
Finally lets save the data dictionary.
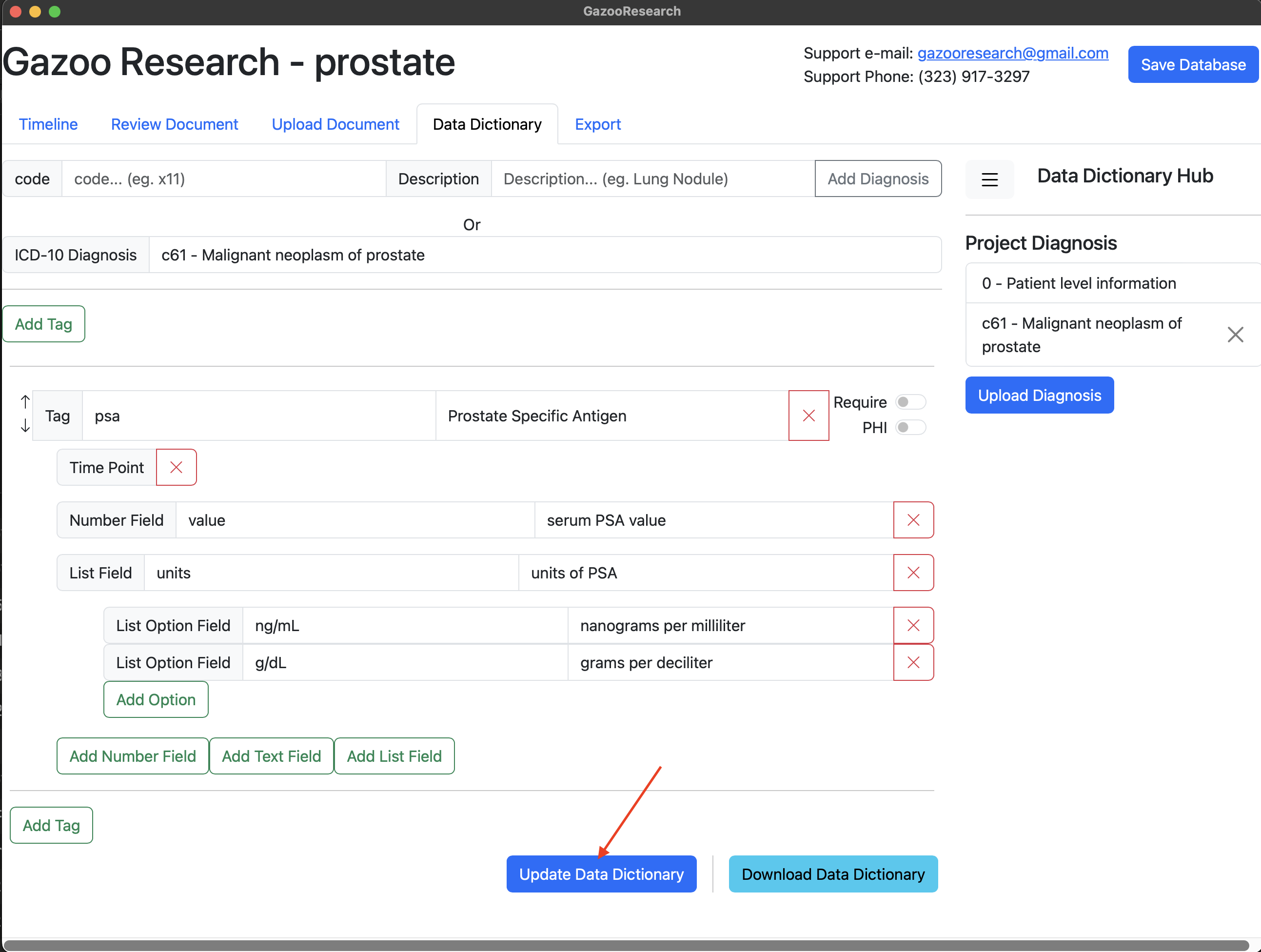 Now when you are annotating documents, you’ll see the “c61:psa” tag
Now when you are annotating documents, you’ll see the “c61:psa” tag
 Additionally, you’ll see the three fields we created: 1) Date, 2) Value, and 3) Units
Additionally, you’ll see the three fields we created: 1) Date, 2) Value, and 3) Units
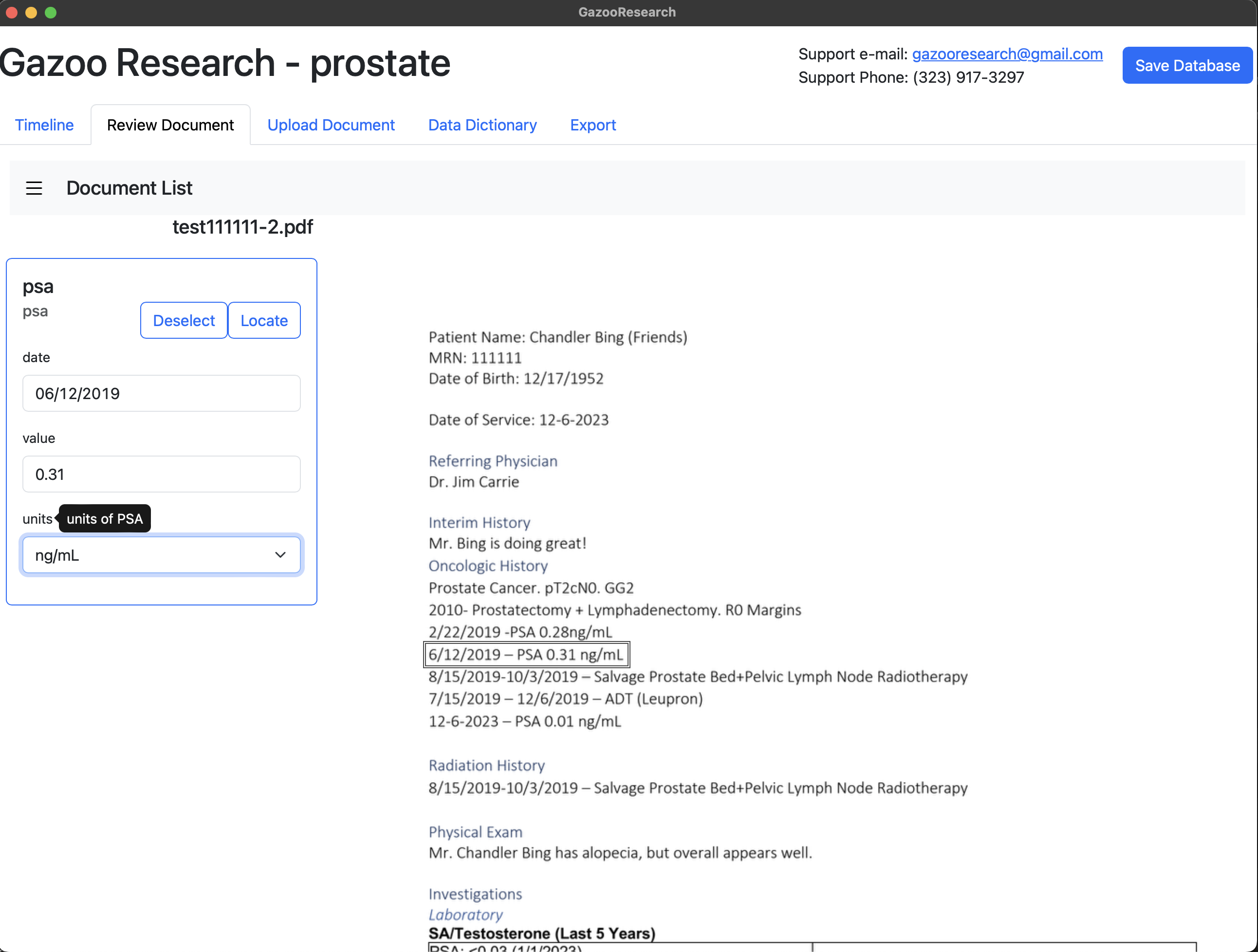
Figure 3. Data Dictionary
Multi-Lesion Data
In some datasets there may be more than one incidence of the cancer, and the researcher may want to analyze the data at the patient level, as well as the lesion level. GazooResearcher supports this type of data aggrigation and analysis.
Examples:
- Skin Cancer: One patient can have multiple independent skin cancers.
- Lung metastasis: One patient can have multiple lung metastasis, each being treated differently (ie. SBRT, Wedge, RFA, Cryotherapy…).
- Prostate Cancer: One prostate may have multiple independent prostate cancer foci, each with different gleason scores.
Example: Multi-Target
In this example we are monitoring lung metastasis size through time. One patient can have multiple lung metastasis.
Data Dictionary
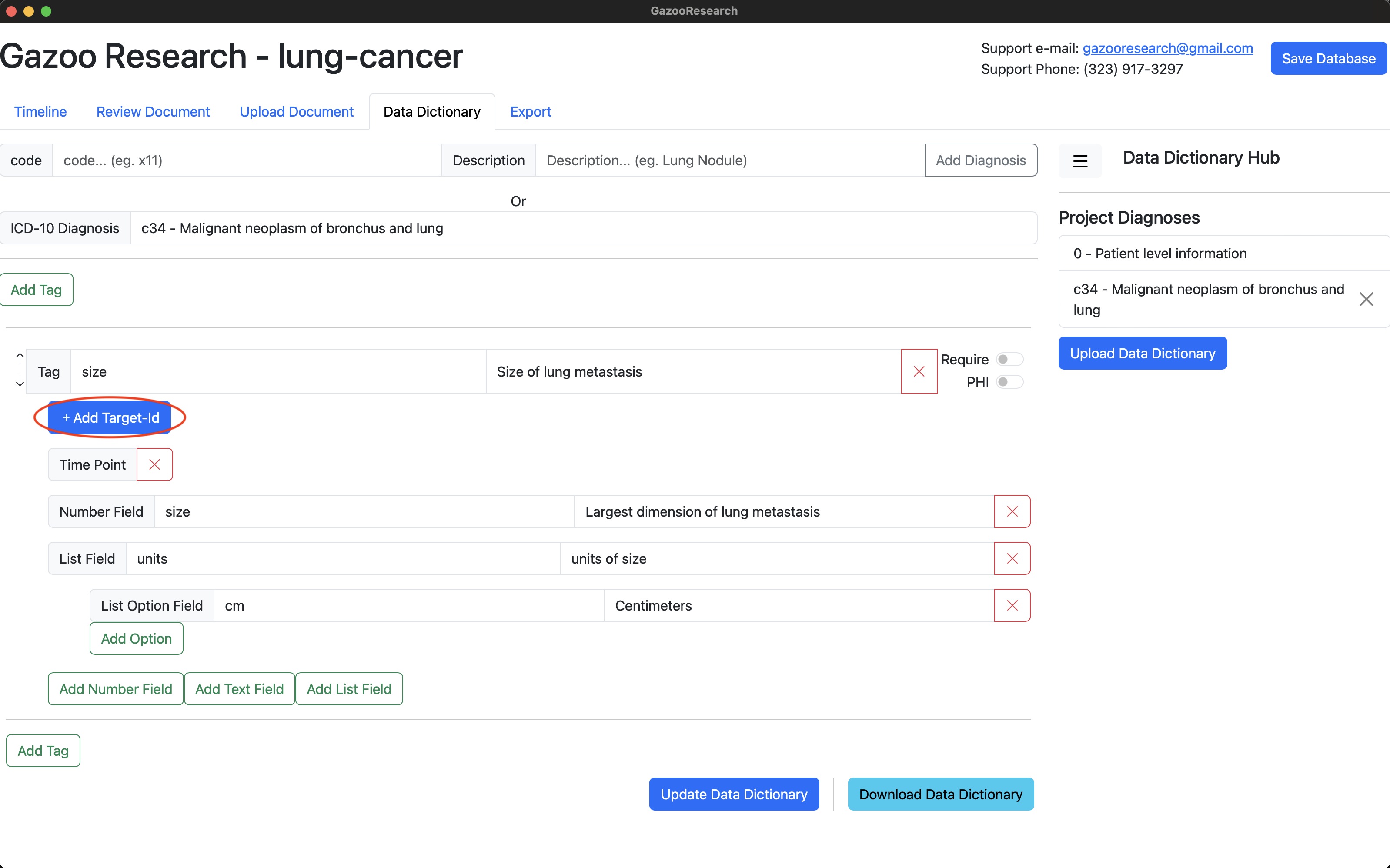
We’ll create a data dictionary using the icd10 code: “c34 - Malignant neoplasm of the bronchus and lung”. The tag will be named “size” indicating the size of the lung nodules. The tag’s fields include 1) Date 2) Size 3) Units, 4) target-id.
Click the “Add Target-Id” button to create a special field named: “target-id”, and add the description
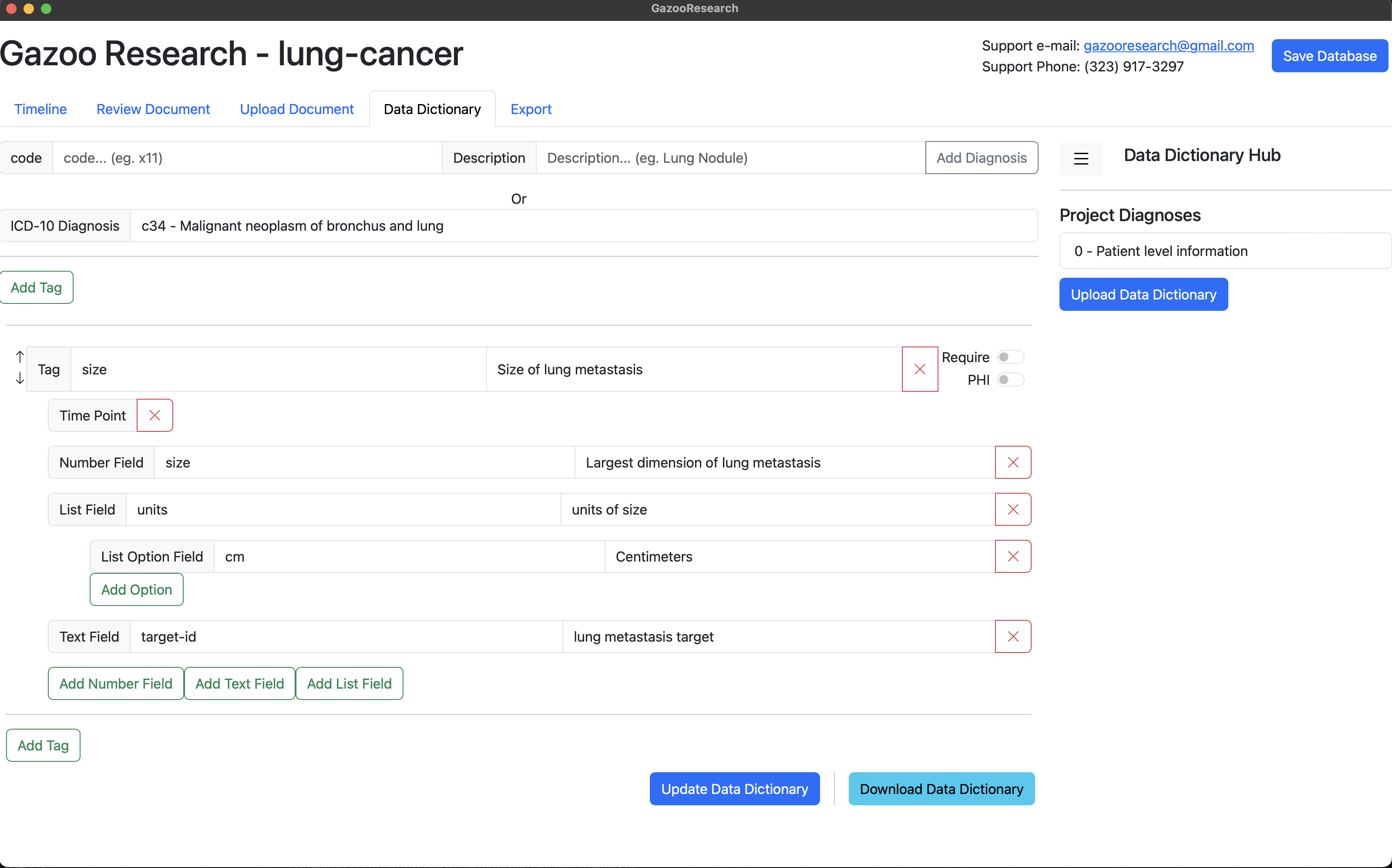
Annotate Data
In the background we have uploaded 3 very basic CT scan reports, and annotated basic information like patient id (ie. mrn), and document.
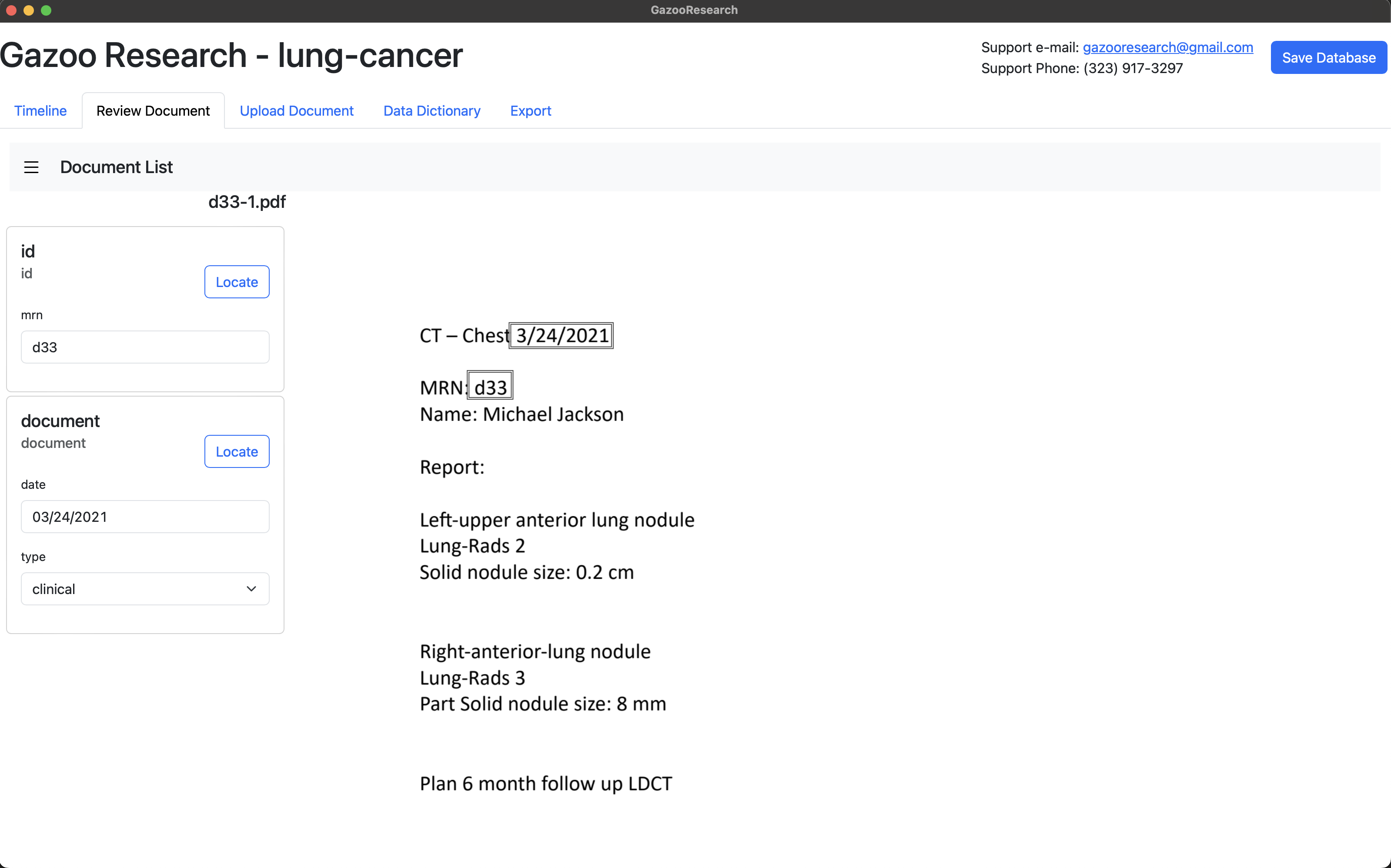
Start annotating the size tag in the documents.
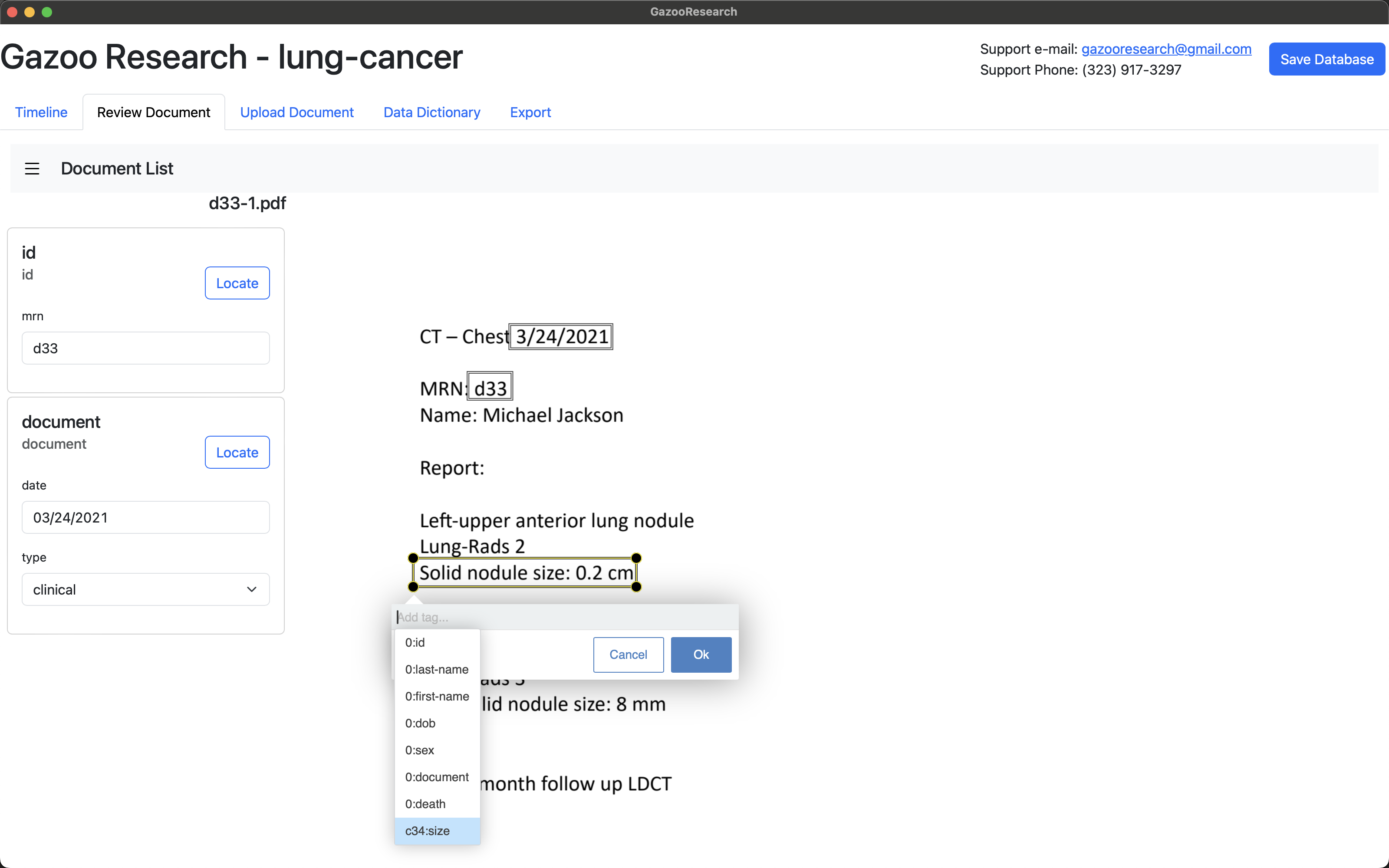 Under the “target-id” field, create a unique name for that target. We’ll use “lul-ant”, which stands for “Left Upper Lobe - Anterior Segment”.
Under the “target-id” field, create a unique name for that target. We’ll use “lul-ant”, which stands for “Left Upper Lobe - Anterior Segment”.
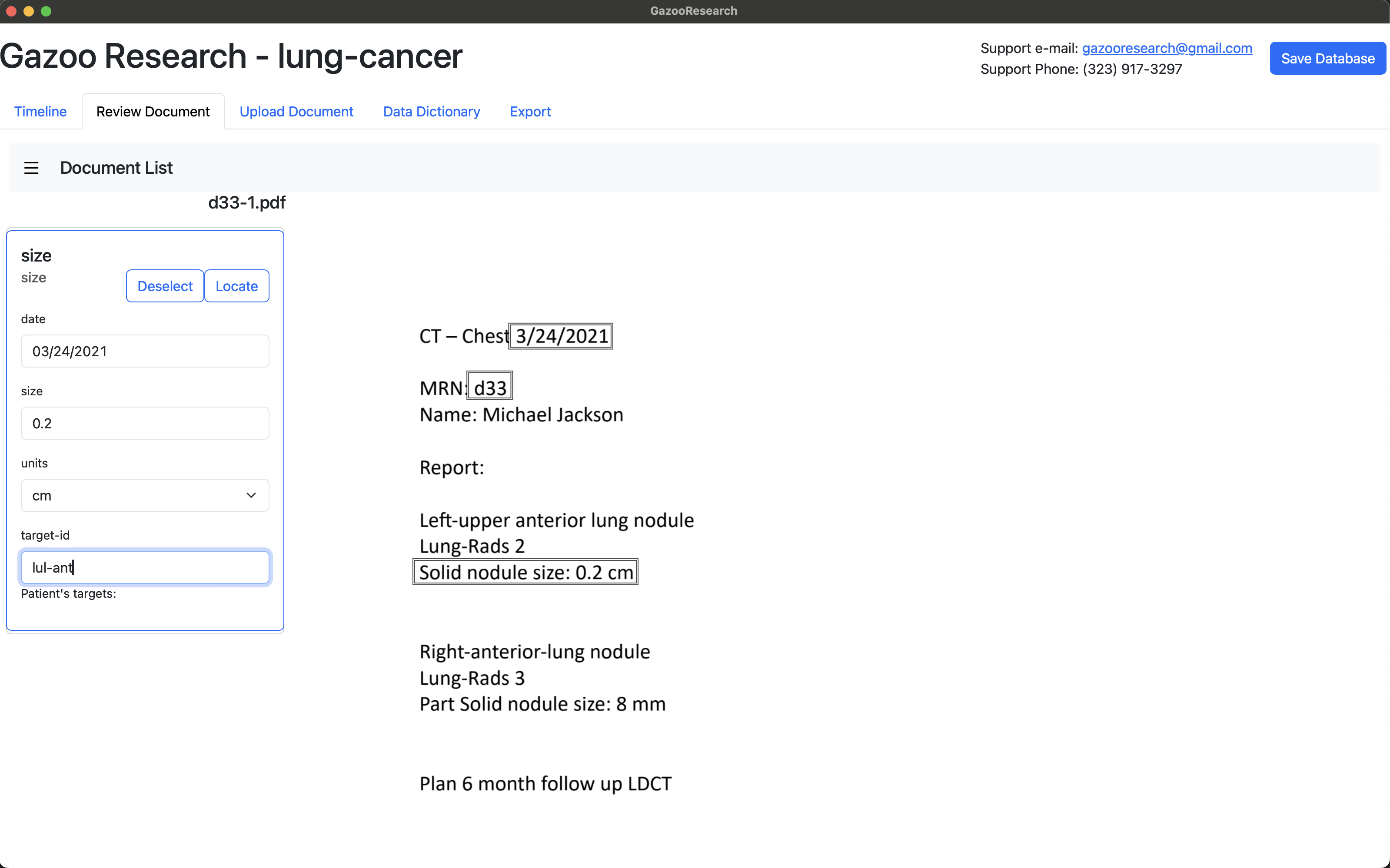 When we annotate the other lung lesion, we’ll use a different “target-id” name: “rul-ant”.
When we annotate the other lung lesion, we’ll use a different “target-id” name: “rul-ant”.
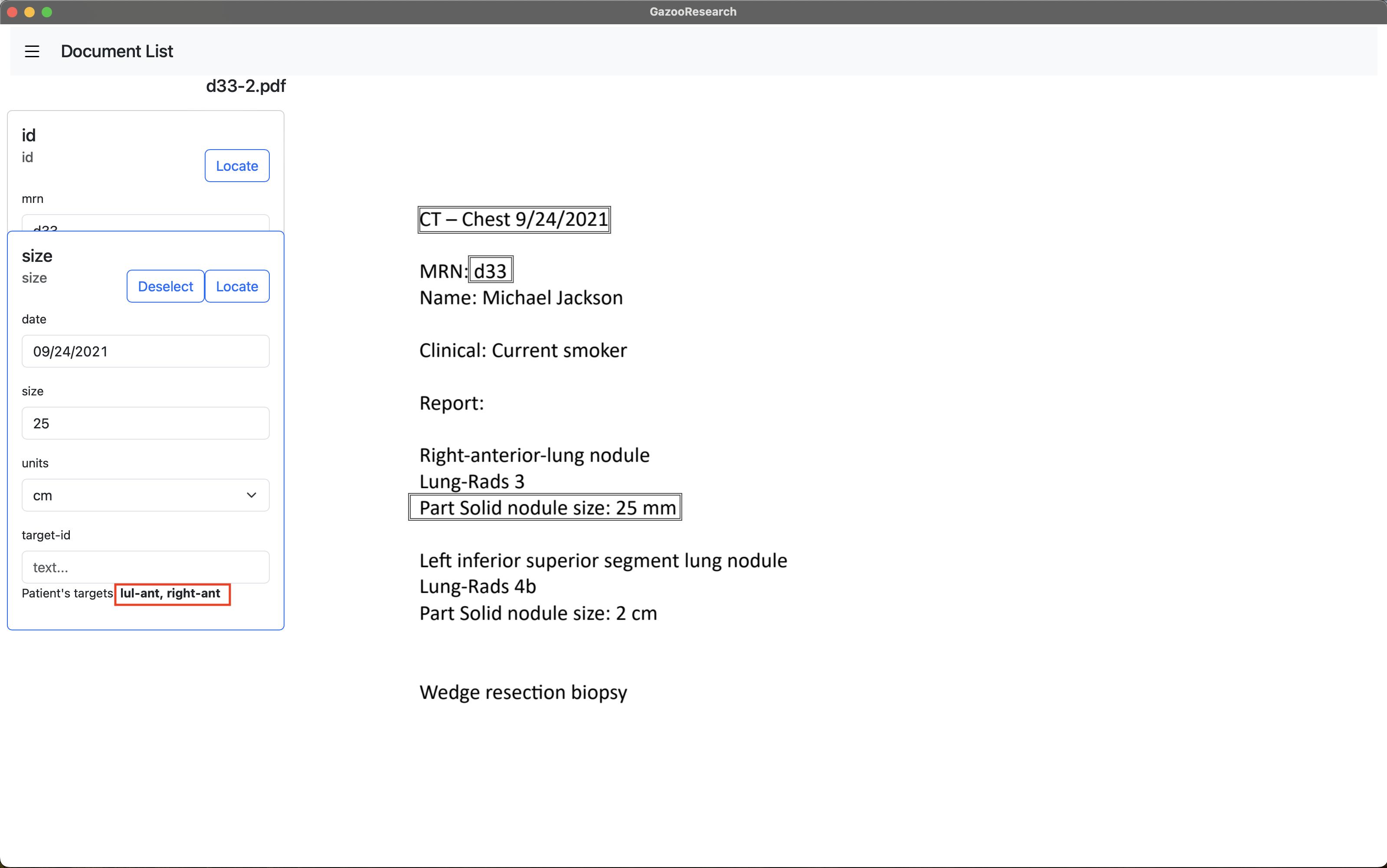
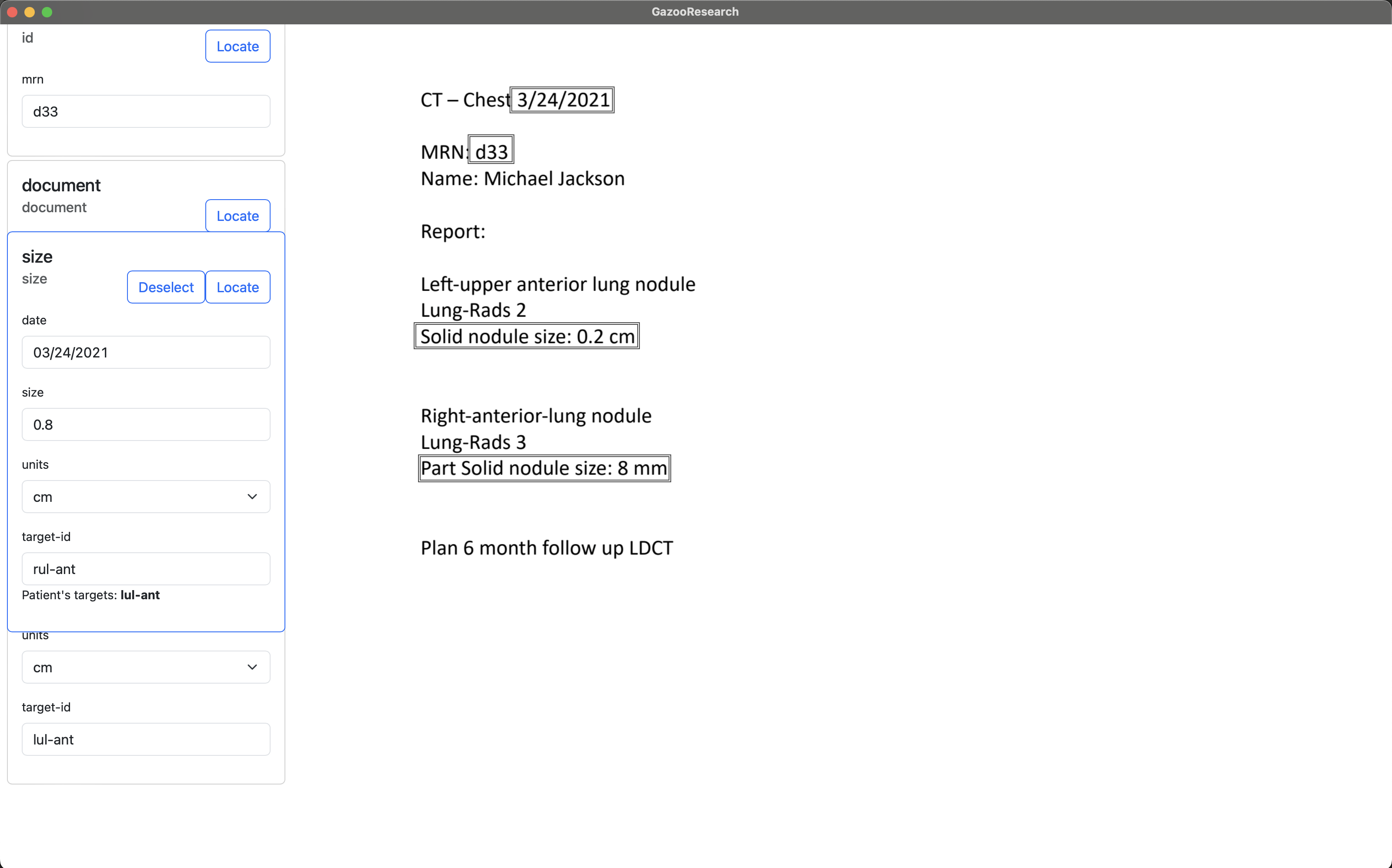 When we navigate to another document, and annotate the tag “size”, we can see that the tag box tells us what target-ids this patient already had. We will enter “rul-ant”.
When we navigate to another document, and annotate the tag “size”, we can see that the tag box tells us what target-ids this patient already had. We will enter “rul-ant”.
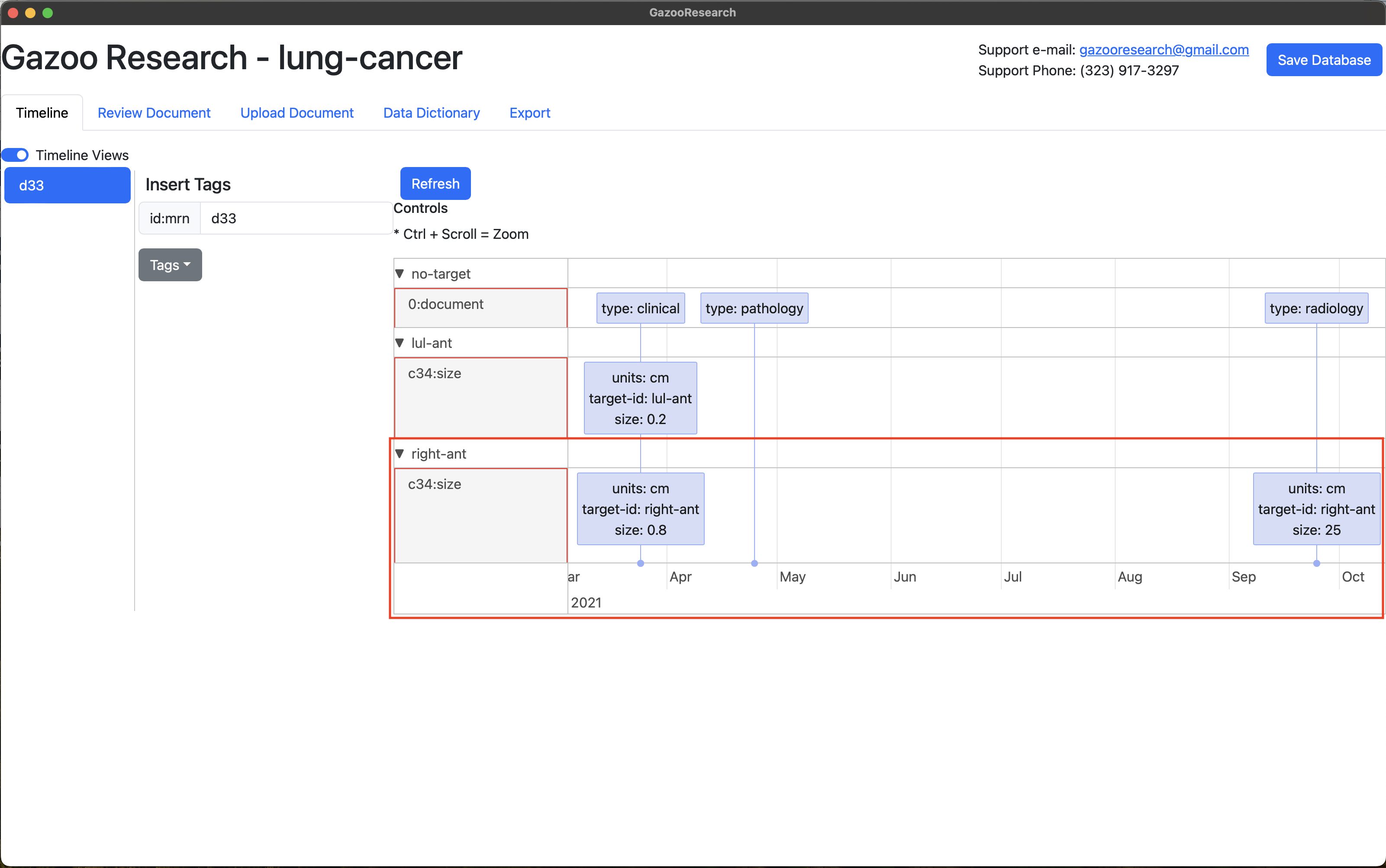
 When you navigate to the Timeline’s Tab. GazooResearch aggrigated target-ids together. You can see that this one lung metastasis grew over the 6 months.
When you navigate to the Timeline’s Tab. GazooResearch aggrigated target-ids together. You can see that this one lung metastasis grew over the 6 months.
Annotation
Data Annotation is a simple as drawing a box around the data you want to highlight, this may be text, an image, or nothing at all.
Steps:
- Draw Bounding Box
- Use the “Down Arrow” to see list of tags.
- Confirm, or modify the extraceted information.
Figure 1. Annotate Data
Timeline
The timelines tab provides a easy way to visualize your database. This is key to ensure data quality. If you believe a data-point is incorrect, you can simply click on the timeline box, and GazooResearch will navigate to where that data was extracted.
Figure 1. Timelines
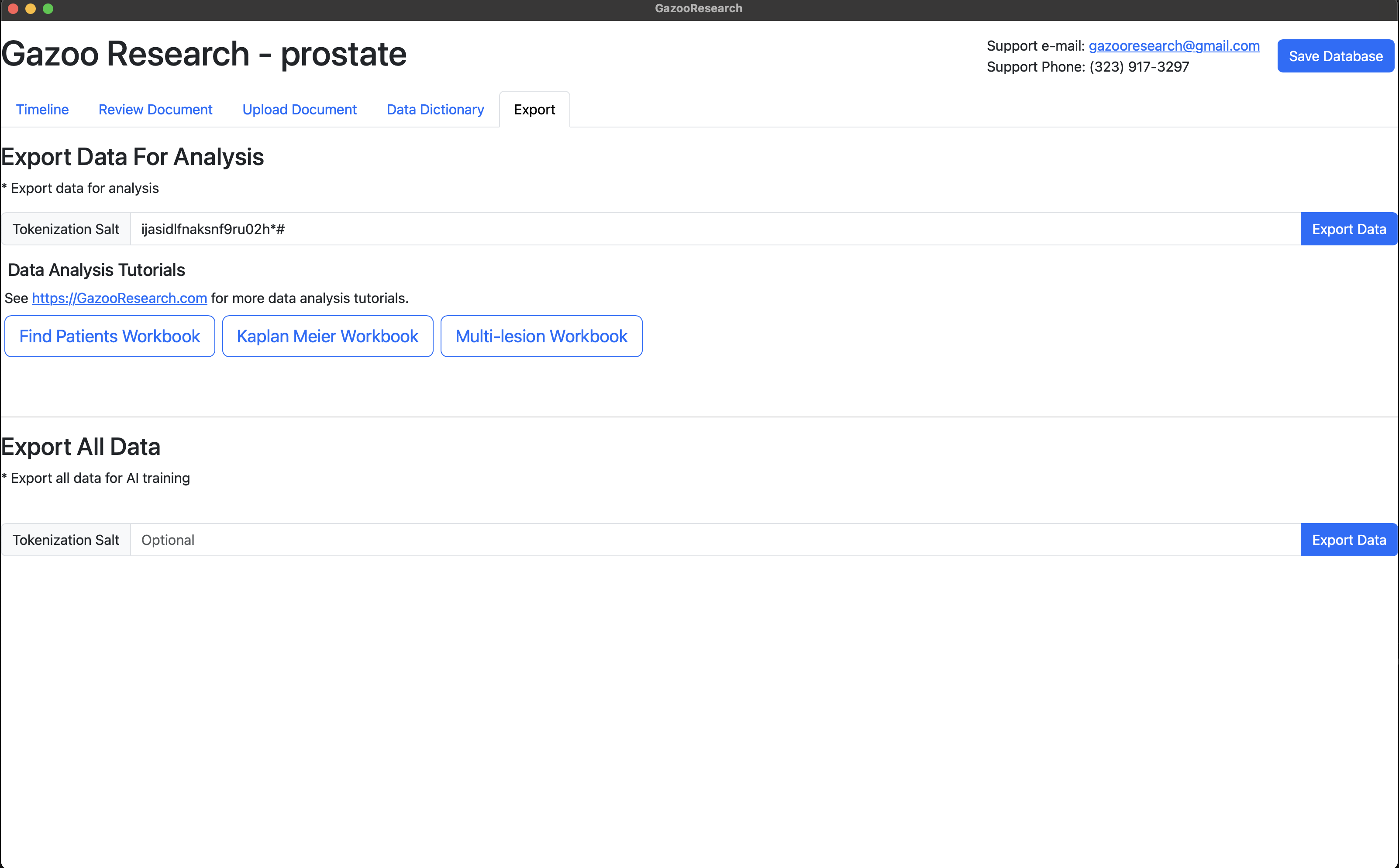
Export Data
Data is exportable for analysis and/or AI training.
The exported date format is: YYYY-MM-DD
Export Data For Analysis
Steps:
- Navigate to the Export Tab.
- Click “Export Data” Button
- Select Save Location and name
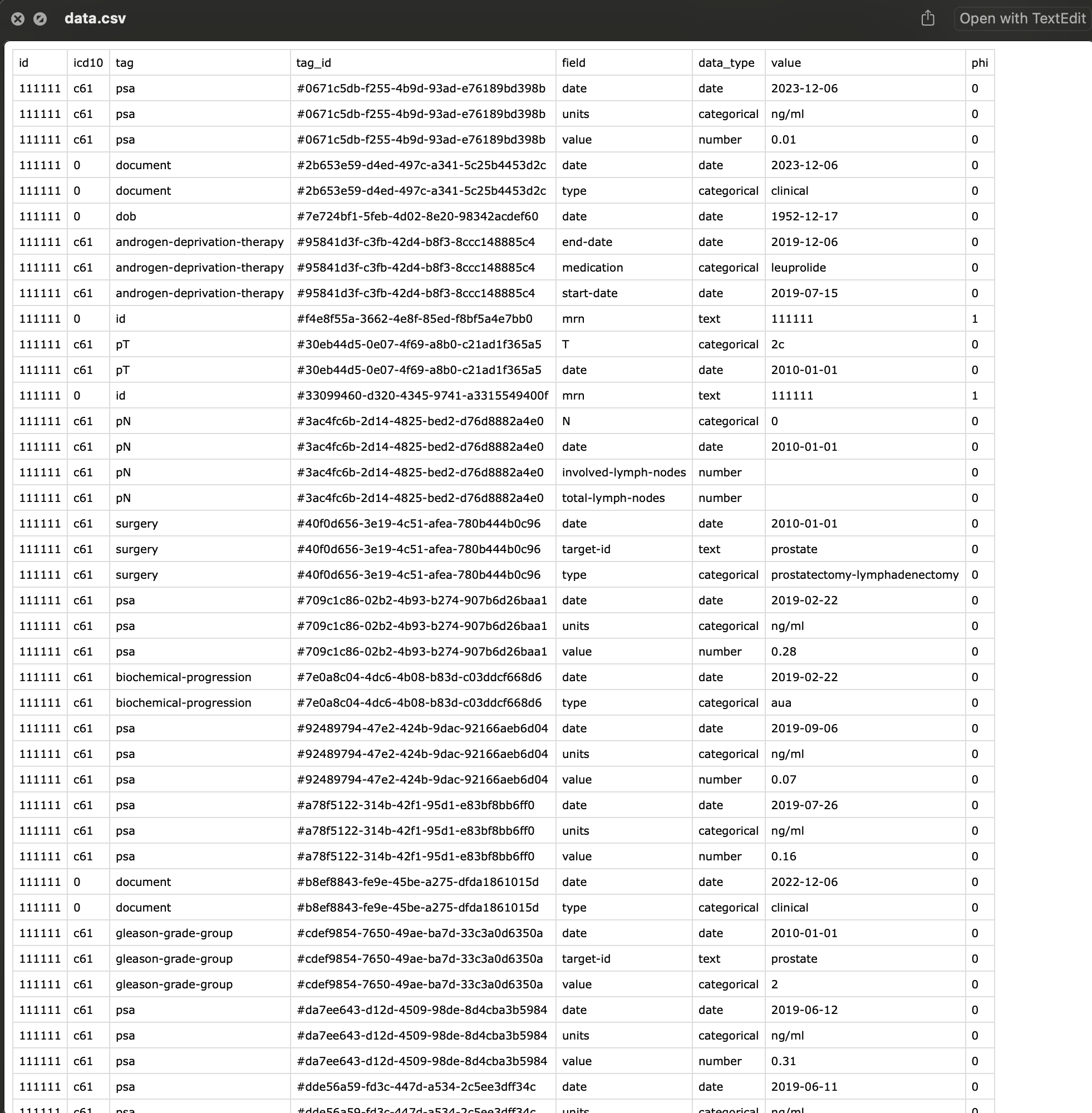
The raw exported data format may not be in the format that you are used to seeing. Don’t worry, we can quickly transform this information into a typical excel spreadsheet in the Analysis section of this book.
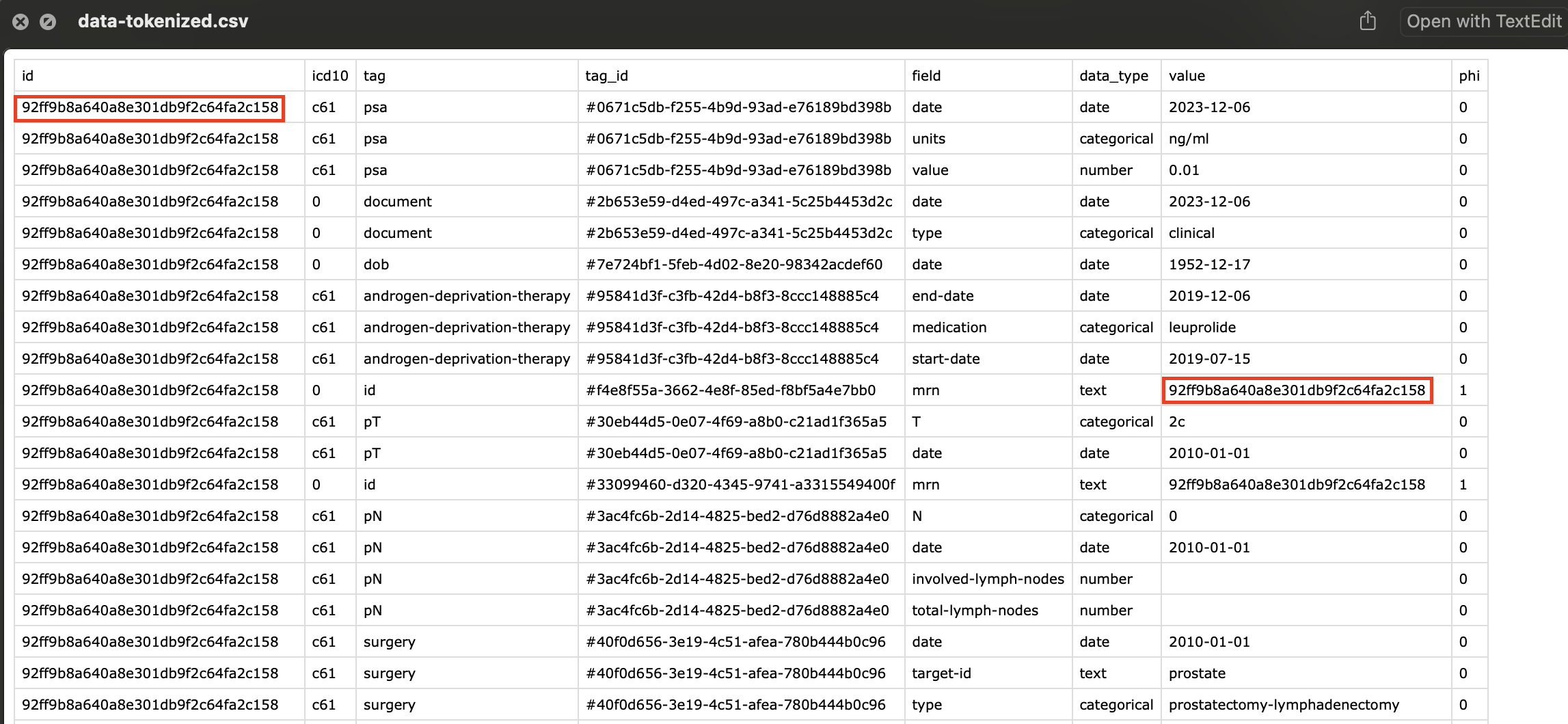
Tokenization
Tokenization is the process of encrypting all the Personal Health Information (PHI) in your data analysis. This is useful if you intend to share your data with other researchers.
If you examine the raw exported data, PHI information is everywhere.
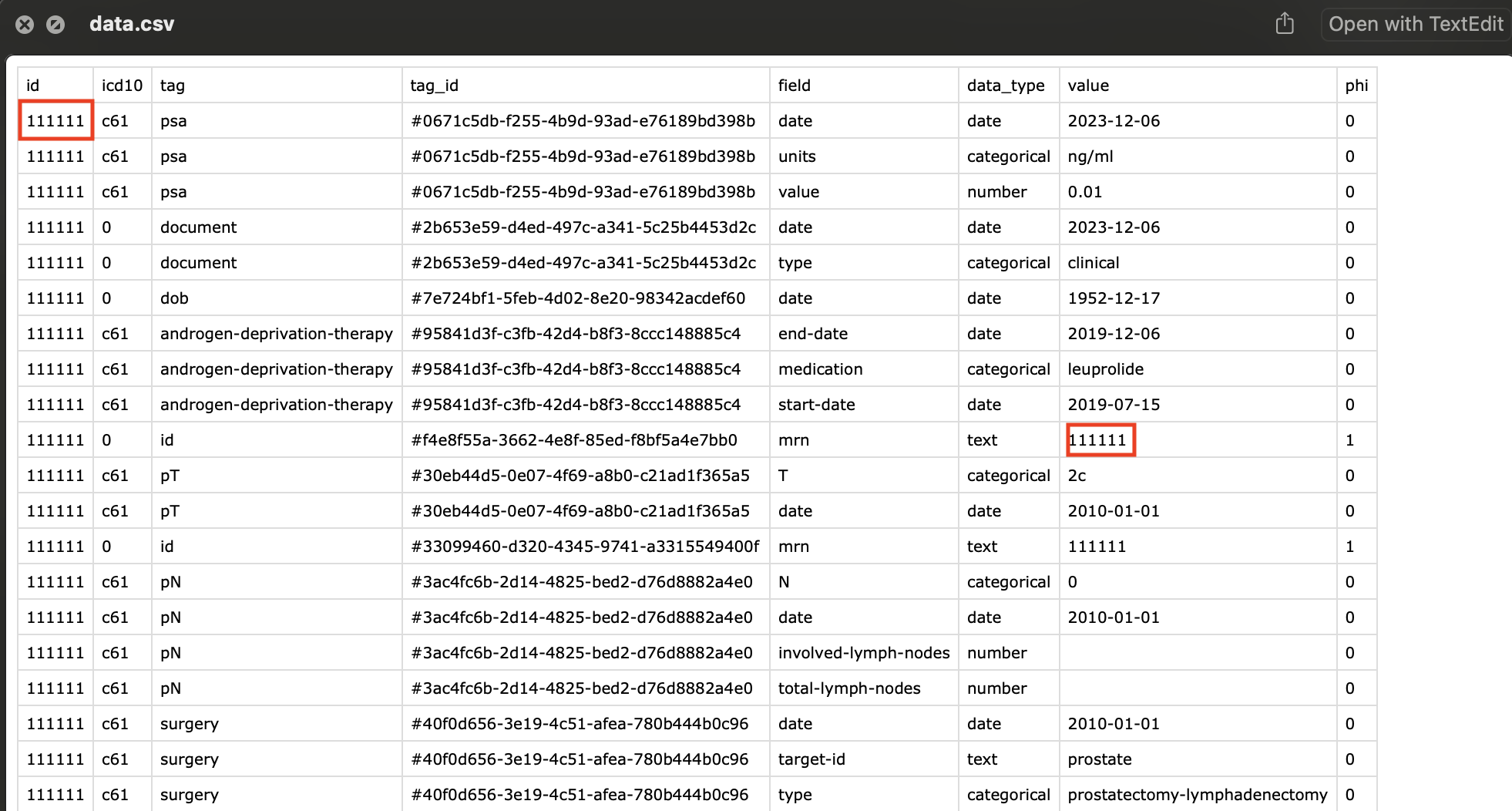 During the export process, if you enter in a random string of characters, this will be used to encrypt all the tags that have been labeled as phi.
During the export process, if you enter in a random string of characters, this will be used to encrypt all the tags that have been labeled as phi.
 All the tags which were labelled as phi have been replaced with an incomprehensible string, preventing anybody from figuring out the original mrn.
All the tags which were labelled as phi have been replaced with an incomprehensible string, preventing anybody from figuring out the original mrn.
Analysis
GazooResearch comes complete with a suite of python based analysis tools, because what is the point of collecting data if you can’t analyse it.
Install and Use GazooResearchUtils Package
!pip install GazooResearchUtils
import GazooResearchUtils as gz
import numpy as np
import pandas as pd
Read Data For Analysis
Upload exported data to Dataframe
df = pd.read_csv("./data.csv")
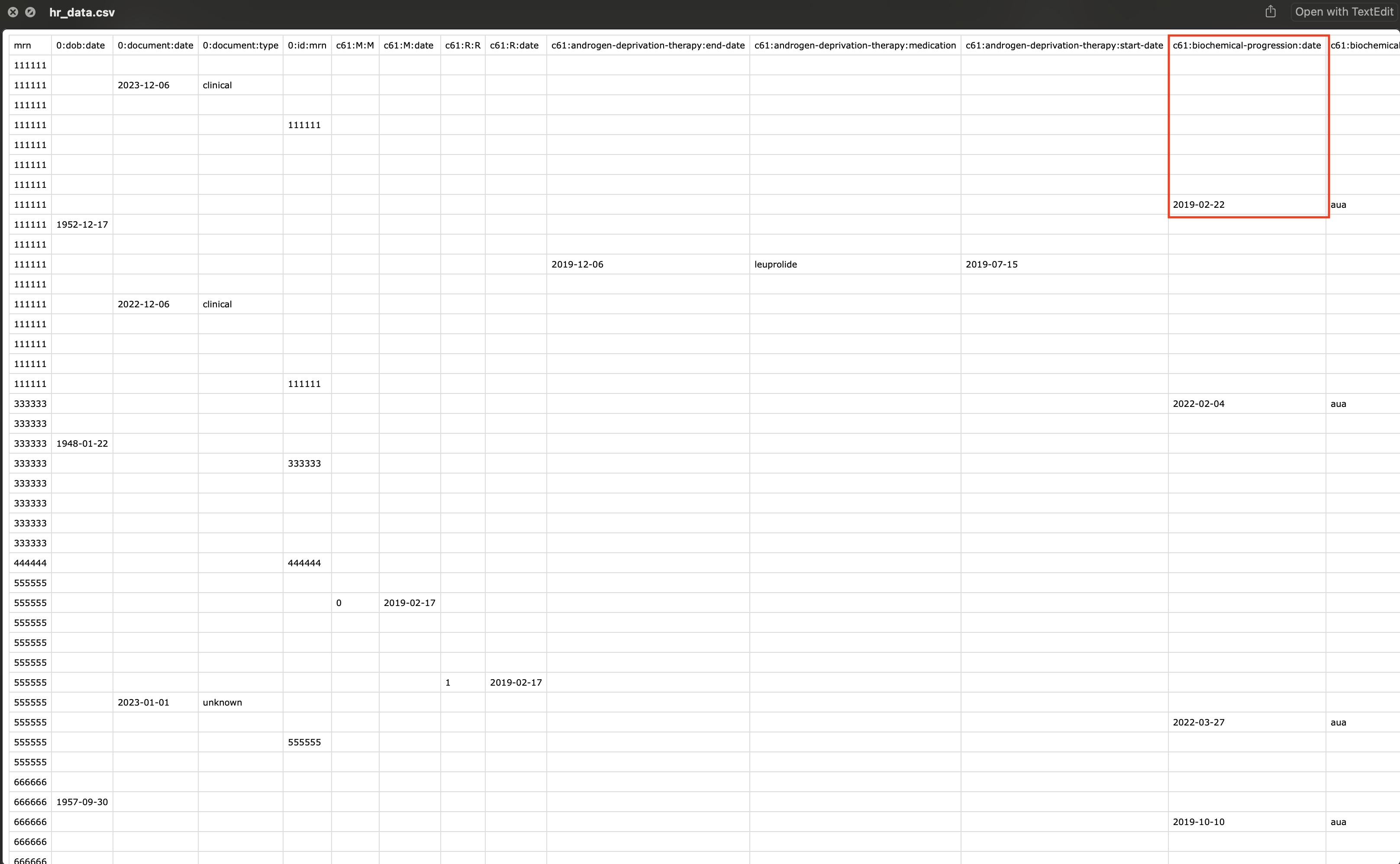
Convert To Human Readable Format
It is recommended to do all the analysis with the default format, and only at the last second, should you convert it to the human readable format.
We use the gz.pivot() function to convert denormalize the data into a more human readable format.
# Convert to human readable format
hr_df = gz.pivot(df)
# Save to csv
hr_df.to_csv("/Users/andrewlim/Desktop/hr_data.csv")
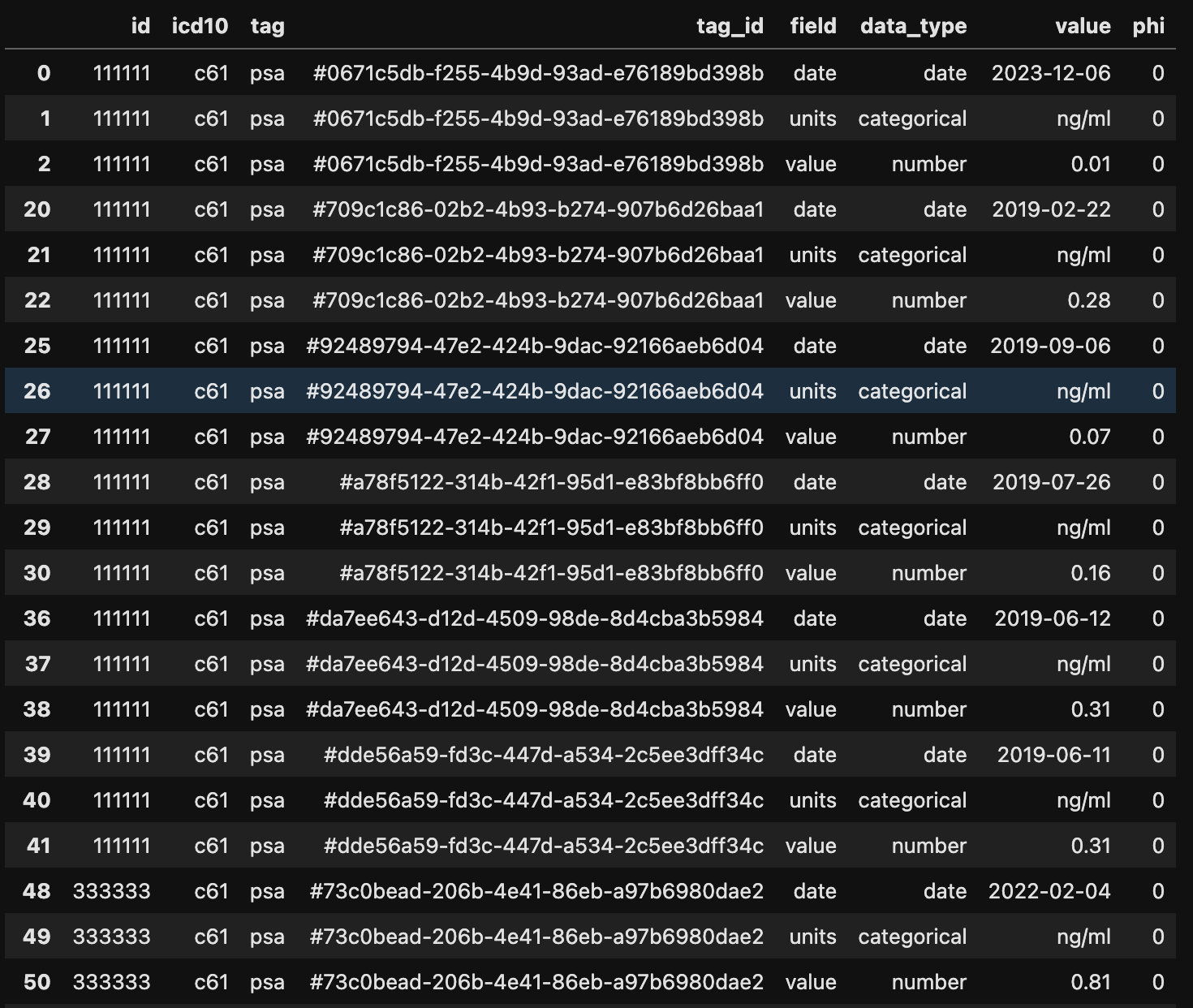
Note that now the column names are the data fields. For example, for mrn:111111, the date of biochemical progression is 2019-02-22.
Plot Data
if you want to plot PSA values, this is what you want to do.
filter = {'tag':'psa'}
gz.get_tags_where_filter(df, filter)
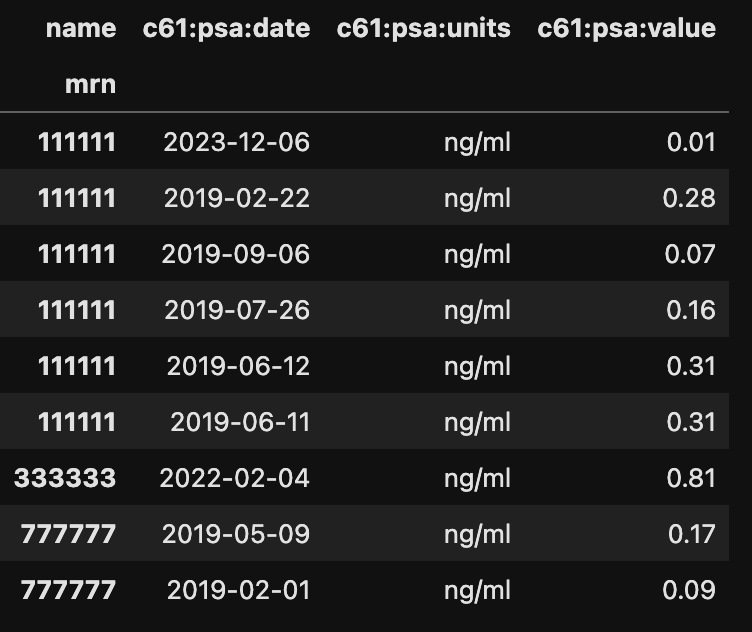
 By simply applying the gz.pivot() function, the data is more readable.
By simply applying the gz.pivot() function, the data is more readable.
filter = {'tag':'psa'}
psa = gz.get_tags_where_filter(df, filter)
gz.pivot(psa)
I recommend being more verbose when specifying the filter object.
# Filter Object Structrue
{'icd10':str, 'tag': str, 'field': str, 'exact': [str, str,...], 'between': [float, float]}
The below finds all the tags which have the psa valube between 0 and 5.
filter = {'icd10':'c61',
'tag':'psa',
'field':'value',
'between':[0.,5.]}
psa = gz.get_tags_where_filter(df, filter)
gz.pivot(psa)
The below finds all the tags which have the pT:T value of ‘2c’, or ‘3a’, or’3b’.
filter = {'icd10':'c61',
'tag':'pT',
'field':'T',
'exact':['2c','3a','3b']}
t_df = gz.get_tags_where_filter(df, filter)
gz.pivot(t_df)
Security
Data security is of the upmost importance for medical information. GazooResearch was developed with data security as priority number 1!
Security Features
On-Premise Design
GazooResearch does not run in the cloud it was designed to run locally on your physical hardware.
*Use of GazooReseach’s AI model(s) requires data to be sent to Gazoo’s servers via encrypted pathways (TLS and other state-of-the-art cryptographic methods), if this is a security issue, then use your OpenAI API compatible LLM servers.
Database Encryption
Gazoo uses an encrypted sqlite database. All the files which make up the database are always encrypted on the disk. It only decrypts blocks as they are read from disk.
Since the data is stored on a disk, we naturally base our approach on “Disk Encryption Theory”. For each type of file, we use the 256-bit AES cipher in the appropriate mode of operation. The AES cipher itself encrypts/decrypts individual files in the most efficient way possible. Your data will be safe on disk.
Document Encryption
Documents are stored on disk using a 256-bit AES CBC mode cipher. 256-bit AES encryption is considered safe against brute-force attacks. It has 2128 potential key choices, making it difficult to crack. A machine that can crack a DES key in a second would take 149 trillion years to crack a 128-bit AES key.
Transport Encryption
Communication between the different components of the software are secure, having been reviewed by a third party.
Suggested Security Features
Full Disk Encryption
MacOS: It’s suggested that you use FileVault to encrypt all data written to disk. Debian: It’s suggested that you use Linux Unified Key Setup (LUKS) hardrive encryption.
Air-Gapped Environment
For further data protection, Gazoo can run in an air-gapped environment (not connected to the internet), this is the gold standard for data security.
Example Security Paragraph
Medical information is secured using 6 layers of security:
Physical data security begins with the medical data being located 1) on-premise, 2) behind physical locked doors. The computer is 3) air-gapped from the outside network, and only accessed physically, with the 4)correct login credentials. The hardrive containing the data is 5)fully encrpyted at rest using the Linux Unified Key Setup (LUKS) which is a trusted hardrive encryption technique. While the computer is turned on, but the medical information is not being accessed (data is ‘at rest’), the 6) data is encrypted using a 256-bit AES cipher.
Format Converter
Gazoo Research accepts pdf, jpeg, png file formats, but does not accept docx, txt files. The gazoo-research-docker docker image can convert all your documents to pdf files so that they can be uploaded to GazooResearch.
Requirements
The Docker app is required.
Usage
You must first create an output directory, which will be where the resultant pdfs will be placed.
-v: mount the input directory to /home/src
-v: mount the output directory to /home/out
docker run -it --rm \
-v <input directory path >:/home/src \
-v <output directory path>:/home/out \
andrewlimmer/gazoo-research-converter
Example
docker run -it --rm \
-v /Users/Desktop/research-data:/home/src \
-v /Users/Desktop/research-data-converted:/home/out \
andrewlimmer/gazoo-research-converter